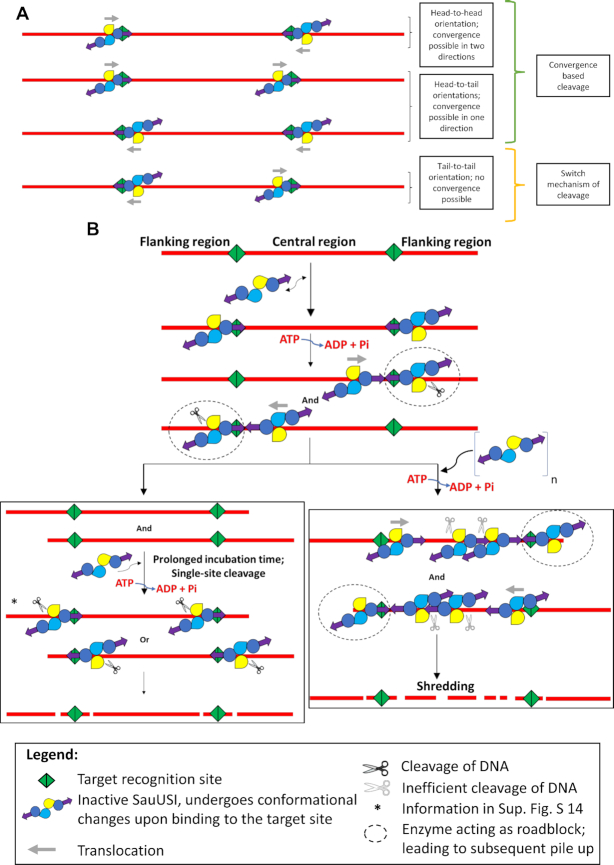
Figure 8.
Model for the cleavage by SauUSI. (A) The different orientations that SauUSI can bind when there are two target sites. The head-to-head and head-to-tail orientations lead to possible convergence events leading to DNA cleavage. In the tail-to-tail orientation, however, there is no convergence event possible; therefore, it is cleaved by the switch mechanism (similar to single-site cleavage). Even though the probability of binding to each of these orientations is equal, cleavage seems to occur more rapidly when there is a possibility of enzymes converging. (B) A cartoon representing the model of cleavage of a two-site substrate when SauUSI molecules are bound in the head-to-head orientation. SauUSI is a homodimer with the interface at the nuclease domain (yellow and blue colors representing the nuclease domains). Each monomer has one helicase domain (dark blue circle) and one TRD (purple arrow). The enzyme binds to the DNA (red) possessing the target sites (green) undergoing a conformational change (represented by a slight change in the orientation in the cartoon). Upon ATP hydrolysis, a translocating SauUSI can converge with a stationary SauUSI (roadblock; black dotted circle around the roadblock) bound to another target site. The convergence stalls the translocating enzyme, and DNA is cleaved by the stationary enzyme (black pair of scissors). There are two routes that the cleavage can proceed depending on the concentration of the enzyme. When SauUSI is present in a limited concentration, the preceding convergence event leads to cleavage producing two two-site fragments. These fragments, upon prolonged incubation with SauUSI and ATP, undergo cleavage due to the switch activity of the ATPase eventually leading to discreet bands. However, when SauUSI is present in excess, then, there is multiple binding and translocation of SauUSI in either direction leading to a pile-up of enzymes along the central region. This, in turn, leads to the stalling of multiple enzymes in the central region. Inefficient nucleolytic activity (grey pair of scissors) of the stalled enzymes leads to random dsDNA breaks, thus shredding the central DNA fragment. Starred steps represent single-site cleavage events (refer to Supplementary Figure S13 for possible products formed).