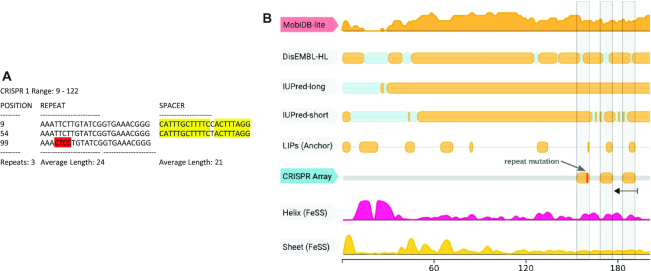
Figure 2.
Example of a spurious detected array. (A) The repeat spacer structure is special in two aspects. First, the two spacers are identical (marked in yellow) with the exception of one position. Second, the third repeat has several mismatches (marked in red). (B) The locus is overlapping a predicted protein on the opposite strand, which has an intrinsically disordered region predicted by several tools to overlap the array position. Furthermore, the three repeats overall exactly with three linear interacting peptides (LIPs) predicted by Anchor. In addition, the first LIP is terminated at the position where the last repeat (in opposite direction) has a mutated region. This together provides a repetitive protein sequence or a duplication event as an alternative explanation for the repetitive structure predicted as CRISPR array.