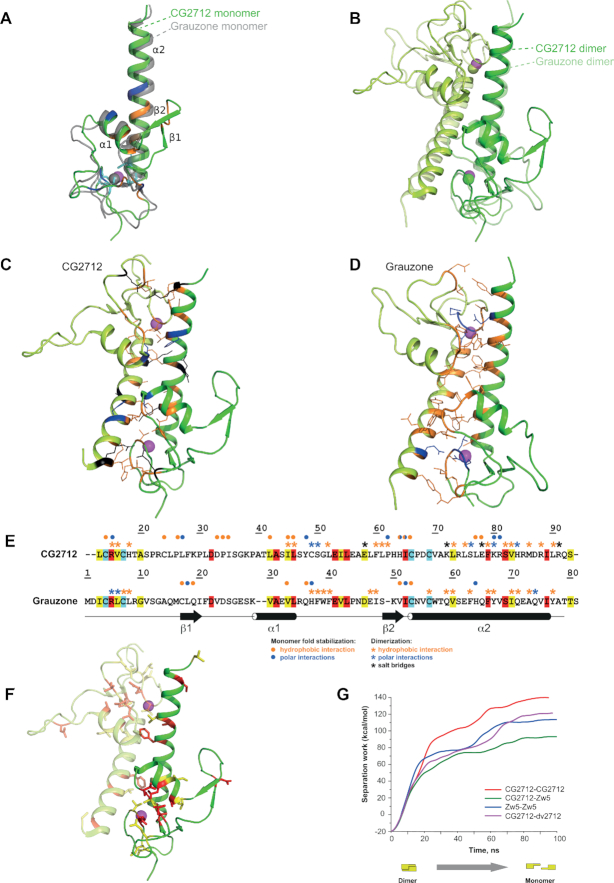
Figure 3.
Crystal structure of CG2712 ZAD is highly similar to that of Grauzone ZAD. (A) The crystal structure of CG2712 ZAD monomer. Residues stabilizing the monomer are colored blue for hydrogen bonds, orange for hydrophobic interactions and cyan for invariant cysteine residues. The zinc ion is colored in magenta. The superposed Grauzone monomer is colored in gray and is semi-transparent for clarity. (B) Overlay of CG2712 and Grauzone ZAD homodimers. Superposition was made using one of the monomers. Dimers are colored by chain, with the Grauzone dimer depicted as semi-transparent. Zinc ions are in magenta and green for CG2712 and Grauzone ZADs, accordingly. (C) Dimeric interface of the CG2712 ZAD homodimer. Side chains of the residues involved in the interface are shown as wire and are colored as blue for hydrogen bonds, black for salt bridges, and orange for hydrophobic interactions. (D) Dimeric interface of Grauzone ZAD homodimer. Color scheme and orientation of the molecule is similar to panel C. (E) Amino acid alignment of Grauzone and CG2712 ZADs (color scheme is similar to Figure 2A). Blue asterisks show residues involved in polar interactions between monomers, black – residues involved in salt bridges, while orange asterisks mark residues forming hydrophobic contacts according to molecular dynamics simulation (CG2712) or crystal structure analysis (Grauzone). Circles show interactions stabilizing the corresponding monomer. Color scheme is the same as for asterisks. (F) Residues conserved between CG2712 and Grauzone ZADs depicted on the crystal structure of CG2712 ZAD. Color scheme is similar to Figure 2A. (G) Results of steered molecular dynamics simulation showing the energy required for dimer dissociation as a function of time.