Abstract
Following antigenic challenge, activated B cells rapidly expand and undergo somatic hypermutation, yielding groups of clonally related B cells with diversified immunoglobulin receptors. Inference of clonal relationships based on the receptor sequence is an essential step in many adaptive immune receptor repertoire sequencing studies. These relationships are typically identified by a multi-step process that involves: (i) grouping sequences based on shared V and J gene assignments, and junction lengths and (ii) clustering these sequences using a junction-based distance. However, this approach is sensitive to the initial gene assignments, which are error-prone, and fails to identify clonal relatives whose junction length has changed through accumulation of indels. Through defining a translation-invariant feature space in which we cluster the sequences, we develop an alignment free clonal identification method that does not require gene assignments and is not restricted to a fixed junction length. This alignment free approach has higher sensitivity compared to a typical junction-based distance method without loss of specificity and PPV. While the alignment free procedure identifies clones that are broadly consistent with the junction-based distance method, it also identifies clones with characteristics (multiple V or J gene assignments or junction lengths) that are not detectable with the junction-based distance method.
INTRODUCTION
A defining property of the adaptive immune system is its capability to adapt to new pathogens. One mechanism underlying this adaptation is the ability to generate B cells expressing a broad range of antibody or Ig receptors and then to modify these receptors upon a antigenic challenge. Each B cell receptor (BCR) is composed of two protein chains, a heavy chain (IgH) and a light chain (IgL). The IgH is created through a somatic recombination process involving a rearrangement of three genes, termed V, D and J, coupled with stochastic insertions and deletions at the gene boundaries (i.e. between the V and D, and the D and J genes). The IgL is generated through a similar process, but without a D gene. This process provides the B cells an initial diversity of ∼107 (1). After a B cell is activated through binding to an antigen and receiving appropriate secondary signals, it can undergo further diversification through somatic hypermutation (SHM), which modifies the BCR mainly through point mutations. B cells with higher affinity to the antigen are preferentially selected to further expand. This process, known as affinity maturation results in a group of clonally related B cells. Identification of these clonally expanded groups are a critical step in the analysis of B cell repertoires. Examples for such biological studies include lineage reconstruction (2–4), diversity analysis (5,6), identification of antigen-specific sequences (7), and more (8,9).
Our ability to analyze large B cell repertoires has improved due to technological advances of Adaptive Immune Receptor Repertoire Sequencing (AIRR-Seq) experiments, which now allow generation of up to hundreds of millions of BCR sequences per sample. Recent studies, e.g. (10–13), use AIRR-Seq to detect properties of the immune system which differentiate between healthy individuals and individuals with cancer, autoimmunity, allergies, or other diseases.
Several methods (14–21) have been proposed to address the challenge of automatic identification of clones from a set of IgH sequences. A common first step is alignment and V(D)J gene identification of each BCR sequence using a reference of known germline V, D and J gene sequences. This step is often performed using IMGT/HighV-QUEST or IgBlast (22,23). Next, sequences are separated into different groups based on shared V and J gene assignments along with identical junction lengths, where the junction is defined as the CDR3 plus the two conserved flanking codons. A distance (indicating a level of similarity) is computed between junctions in these smaller groups, and some form of clustering is then used to identify the clones. Various distance metrics have been used to compare the junctions, including a Hamming distance (14,15), the Levenshtein distance (16,24) and metrics which incorporate SHM hot- and cold-spot motifs (21,25). The Hamming distance is computationally efficient, but restricted to fixed sequence length comparison. The Levenshtein distance removes this restriction, but with a high computational cost and therefor does not scale to huge repertoires. Furthermore, as reported in (16) the Levensthein distance is sensitive to insertion and deletions and obtains PPV values 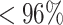 (even when incorporating gene assignments).
(even when incorporating gene assignments).
To distinguish between clonally related and unrelated sequences, earlier studies set a fixed threshold on the distance between junctions (26–28). The authors in (17) noticed that the distribution of distances between sequences and their nearest neighbors (distance-to-nearest) tends to be bi-modal, with a first mode corresponding to clonally related sequences and second mode corresponding to sequences without clonal relationship (singletons). Using this bi-modality, (17) proposes to set a threshold that separates the two modes. Following this observation (14,20), use the bi-modality of this distribution to suggest an automatic way to set the threshold. A recent method by (15) uses spectral clustering with an adaptive threshold to identify the groups of clonally related sequences.
Methods such as (14–15,20) identify clonally related sequences with high confidence (29); however, their success relies on two assumptions, namely that all clone members should share the same V and J gene assignments, as well as a common junction length. The later assumption effectively ignores the possibility of SHM to introduce insertions and deletions (indels). The former premise relies on the success of a pre-processing method that aligns the sequences and assigns the V and J genes. Although most alignment methods use similar germline gene databases (such as IMGT) the gene assignments may only partly overlap (30). Even for a low mutation rate of 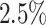 , the assignment errors of the V and J genes can be
, the assignment errors of the V and J genes can be 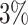 (31). Subsequently, these two types of errors will lead to a non-negligible number of incorrect clonal assignments.
(31). Subsequently, these two types of errors will lead to a non-negligible number of incorrect clonal assignments.
In this study, we present an alignment free approach for clonal identification; this enables us to bypass the V(D)J gene assignment step and remove the fixed junction length restriction. The alignment free method is based on techniques from natural language processing (NLP), specifically, we use k-mer representations and re-weight them with a term frequency inverse document frequency (tf-idf). The tf-idf is a statistical measure widely used in NLP. Next, by applying a cosine distance to the re-weighted representation, we define a tf-idf distance, which allows us to identify sequences derived from clonally related B cells. This procedure does not require sequence alignment and is not restricted to sequences with the same junction length. In Figure 1, we illustrate how the tf-idf distance bypasses three building blocks from the standard distance-based clonal assignment procedure. In the Materials and Methods section, we describe the proposed alignment free approach, and detail alternative approaches for clonal assignments. In the Results section, we evaluate the capabilities of the alignment free methods using artificial and real repertoires.
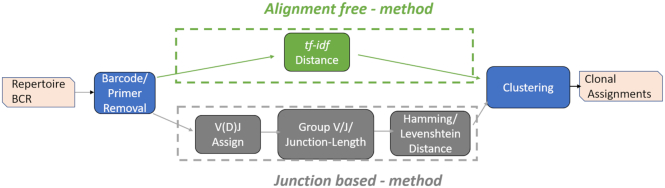
Figure 1.
A flow diagram depicting the major steps for identifying clonally-related B cell receptor sequences (bottom row). Given a set of BCR sequences (the repertoire), first, the primers and barcodes are removed, then V(D)J genes are assigned based on an alignment of the sequences to a database of germline genes. Sequences are grouped based on V and J gene assignments and junction length. A hamming distance is calculated on the junction regions of pairs of sequences in each of the groups separately. Finally, distances are fed into a clustering algorithm (Hierarchical (14) or Spectral (15)). Here, we propose to use a tf-idf based distance that bypasses the three steps prior to clustering, and is not restricted to sequences with the same V or J gene or junction length.
MATERIALS AND METHODS
In this section, we describe the proposed alignment free approach, as well as an alternative method providing a baseline.
Junction-based clonal identification
As a baseline, we compare the performance of the proposed method to the fixed threshold, clustering-based approach proposed by (14), which has been shown to identify clonal relationships with high confidence. This junction-based approach first separates the BCR sequences into different groups that share the same V and J gene annotations, as well as a common junction length. Then, the Hamming distance is computed between all pairs of junctions from the same group, and the distribution of nearest neighbors for each sequence (also termed distance-to-nearest distribution) is analyzed to find a fixed distance threshold for single-linkage hierarchical clustering. This distance-to-nearest distribution is often bi-modal (e.g. see Figure 2), where the first mode is assumed to correspond to distances between members of the same clone and the second mode to distances between sequences from different clones. The method identifies clonally-related sequences by aggregating sequences that share a nearest neighbor distance smaller than the value (threshold) that separates the two modes of the distribution (1). The threshold is determined using a two-step process. First, the distribution of the distance-to-nearest is computed using a kernel density estimator. Next, the threshold is estimated as the first local minimum by calculating the first and second derivatives of the density estimate. As shown in (20), this is computationally expensive and empirically scales exponentially with the number of sequences. To improve run time, (20) fit a Gamma and a Gaussian distribution to the bi-modal distribution and use Maximum Likelihood to determine the threshold. This method seems to scale linearly with the number of sequences. Besides the improvement in run time, both approaches fail if the distribution is not bi-modal. This occurs if there are few or no singletons in the data, i.e. most sequences are members of clones of a size larger than one. A method to overcome this caveat was presented in (2); the authors use spectral clustering, which allows them to identify the clones without restricting to a single threshold per repertoire.
Figure 2.
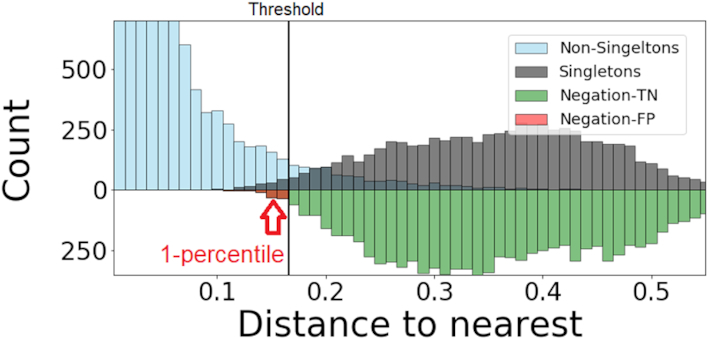
An example of a distance-to-nearest distribution based on an artificially generated repertoire, so that all clonal relationships are known. The bi-modality of the distribution is evident. Blue bars correspond to sequences that belong to a clone (non singletons), while gray bars represent sequences with no clonal relatives in the data set (singletons). Green bars represent the distribution of distances to closest sequences pooled from alternative individuals (negation sequences). The distance threshold (vertical solid line) is set as the lowest 1 percentile of the negation distribution. This values aims for 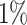 of false positives (FP) and
of false positives (FP) and 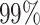 of true negatives (TN).
of true negatives (TN).
Throughout the experiments presented in the Results section, in order to use the junction-based distance method, we first apply IgBLAST with the IMGT gene references to infer gene segments and the junction location. Then, we use the Change-O-DefineClone function from SHazaM R package (version 0.1.11) (32) to define the clones.
k-mer representation of terms
A k-mer representation maps a sequence of length L to the set of all possible sub-sequences of length k. k-mers have been widely used in the context of DNA analysis due to their ability to reduce the computational effort of comparing DNA strings. The k-mer representation of a sequence is a vector with 4k entries, where each entry corresponds to the number of times a specific k-mer is detected along the sequence using a sliding window scheme. This construction ignores the locations of particular k-mers within each sequence.
tf-idf representation
A challenging task in natural language processing (NLP) involves comparing documents with a large and varying number of terms. One popular approach involves using a term frequency inverse document frequency (tf-idf) (33). The tf-idf weighting scheme aims to emphasize the rare and hopefully meaningful terms and reduce the influence of common terms. The tf counts the number of term appearances in the document, while idf measures the importance of the term by counting its appearances in the corpus. A number variants of the tf-idf have been proposed in the literature, for examples see (34,35).
In this study we adapt the tf-idf to reweigh k-mer based representation of BCR sequences. The term-frequency tfs(k) is a count table of the amount of k-mers k ∈ K present in each sequence s ∈ S. The tf is reweighed using the inverse document frequency which is defined as 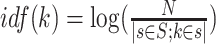 , where N is the total number of sequences and the denominator is the total occurrences of specific k-mer k across all the S sequences. The tf-idf is then defined as tf − idfs(k) = tfs(k) · idf(k).
, where N is the total number of sequences and the denominator is the total occurrences of specific k-mer k across all the S sequences. The tf-idf is then defined as tf − idfs(k) = tfs(k) · idf(k).
Fast cosine distance
To compare the tf-idf representations of sequences s and s′ we choose the widely used cosine distance. The cosine distance is defined as one minus the normalized inner product between two tf-idf vectors, given sequences s and s′ it is computed by
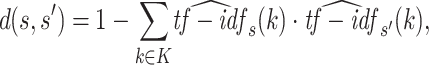 |
where 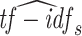 is the L2 normalized tf-idf representation of sequence s. In practice, the tf-idf representation is sparse and clone assignment only requires identification of the most related sequences in terms of proximity (few hundreds, or thousands depending on the size of the clones in the dataset). We can exploit these two properties to reduce the computational cost involved in evaluating the tf-idf cosine distance. The python implementation of the fast cosine distance is available at https://bergvca.github.io/2017/10/14/super-fast-string-matching.html.
is the L2 normalized tf-idf representation of sequence s. In practice, the tf-idf representation is sparse and clone assignment only requires identification of the most related sequences in terms of proximity (few hundreds, or thousands depending on the size of the clones in the dataset). We can exploit these two properties to reduce the computational cost involved in evaluating the tf-idf cosine distance. The python implementation of the fast cosine distance is available at https://bergvca.github.io/2017/10/14/super-fast-string-matching.html.
Automatic clonal distance threshold determination by negation
As in (14,20) we use the properties of the distance to nearest distribution to identify the threshold for cutting the hierarchy of distances. Instead of using the bi-modality, which has a high computational cost, we take a different approach; we propose to find the threshold by negation. The idea is described as follows: given a set of sequences taken from one individual, we introduce a set of sequences randomly sampled from multiple unrelated individuals (negation sequences); then, we compute the distribution of distances between negation sequences and their closest counterpart within the individual. Finally, we set the threshold such that a fraction of the distances to negation sequences that are below the threshold is δ ≥ 0. By definition, clones can not span multiple individuals. Therefore, by choosing a fixed value for δ > 0 (e.g. δ = 0.01) we allow a fraction of false-positive rate roughly equal to δ. This heuristic aims for high specificity, which is approximately 1 − δ. We present an example of such distribution along with the estimated threshold in Figure 2.
Simulation of clonal expansions
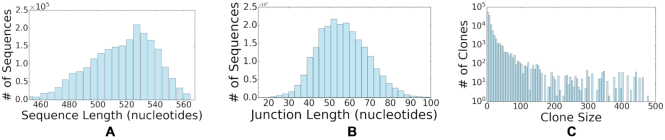
To generate artificial repertoires with known clonal relationships, we first select clone representatives from B cells collected by (28) and filtered to maintain only naive sequences from healthy individuals as in (36). Next, we infer tree topologies of each clone by applying Change-O-buildPhylipLineage (version 0.4.5 (32)) to data from multiple individuals collected in (8). Finally, new artificial samples are generated by randomly adding mutations based on the learned topologies using shmulateTree from the SHazaM R package (version 0.3.0 (32)). We repeat this process using repertoires from four subjects collected in multiple sclerosis (MS) study (37). The corresponding four datasets which contain samples from lymph nodes and are denoted as MS2, MS3, MS4 and MS5 with around 100k, 150k, 200k and 200k sequences, respectively. Using the sequences from the four individuals we generate 74 simulated datasets with ∼30k sequences each. To support the diversity of the artificial datasets, we analyzed the properties of the resulting 74 datasets and observed that the distributions of sequence and junction lengths both follow Gaussian-shaped distributions with means of 521 and 58 nucleotides, respectively. Moreover, as evident in Figure 3, the generated repertoire has a wide range of sequence and junction lengths and diverse clone sizes.
Figure 3.
Statistical properties of the 74 artificially generated datasets. (A) The distribution of sequences length. (B) The distribution of junction lengths. (C) The distribution of clone size (number of unique sequences).
RESULTS
We propose an alignment free method for identification of clonally related sequences. The approach relies on a tf-idf based distance that is invariant to translations of the sequence and is not restricted to the comparison of sequences with the same junction length. The alignment free method does not require alignment of sequences to germline V, D or J genes. The only pre-processing required is removing primers and truncating the sequences to a fixed length from the 3′ end. Here, we evaluate the alignment free approach using both simulated and real datasets. For the simulated repertoires, the correct clonal assignments are known; this allows us to compute the sensitivity, specificity, and positive predictive values (PPV) for nodes and edges, where nodes correspond to sequences and edges correspond to known or inferred clonal relationships. We define node sensitivity as the ratio between sequences identified as part of a multi-sequence (i.e. expanded) clone relative to the total number of sequences which belong to an expanded clone. We calculate node PPV as the ratio between sequences correctly retrieved as part of an expanded clone and the total number of sequences which are assigned to expanded clones. For node specificity, we compute the fraction of sequences identified as singletons relative to the total number of singletons in the repertoire. Next, We define edge sensitivity as the ratio between pairs of sequences (represented by edges) correctly identified as clonally-related and the total number of sequence pairs which truly belong to the same clone. We calculate edge PPV as the ratio between correctly assigned pairs of sequences and the total number of sequence pairs assigned as clonally-related. For edge specificity, we compute the fraction of identified unrelated sequence pairs among all truly unrelated sequence pairs. Illustrative examples for the sensitivity, specificity, and positive predictive values (PPV) computations are presented in Supplementary Figure S6. To further evaluate the alignment free method using real repertoires, we focus on a set of published BCR repertoires from lymph nodes of four multiple sclerosis (MS) subjects referred as MS2-MS5.
idf normalization using a fixed sequence length improves sensitivity and specificity
We start by evaluating how the idf normalization of k-mer representation influences the performance of the alignment free approach. The alignment free method uses nucleotide k-mers reweighed by a tf-idf normalization to find a translation-invariant representation for BCR sequences. We hypothesize that this representation captures sufficient information to allow efficient clonal identification. The term-frequency tfs(k) counts the number of occurrences of each k-mer; thus, the total count of terms per sequence s is affected by the sequence length. As shown in Figure 3(A) the length of the sequences range between 460 and 560 nucleotides. In practice, the bounds of this range can vary depending on the experimental library preparation method although there will always be a distribution of lengths resulting from the V(D)J recombination process. Such variability in the sequence length, in turn, translates to a variability in the amount of k-mers present in each sequence. As shown in (38), such variability may bias the tf-idf based representation. Specifically, applying cosine similarity favors retrieval of short documents (sequences) over long ones.
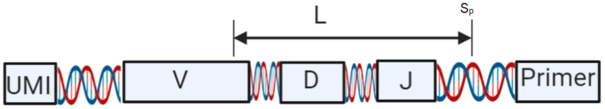
In (38), the authors propose to use a pivoted normalization to compensate for the length effect. Here we can exploit the known structure of the BCR sequences to propose an alternative solution by defining a representation that uses a common number of nucleotides for all sequences. This can be obtained by truncating each sequence using a fixed number of nucleotides (L) starting from the 3′ end. The length of the sequence should be sufficient to cover the J segment, the junction region, and part of the V segment. See Figure 4, in which we illustrate this truncation. We expect that a good truncation should cover the junction region, as it is highly diverse and has been used as a signature for identifying clonally-related BCRs in several studies (14,15).
Figure 4.
Schematic representation of a BCR sequence. The alignment free method uses a fixed number of nucleotides (L) from the 3′ end of the sequence after barcode and primer removal. Different repertoires use different library preparation procedures; thus, the starting position (Sp) may vary and the sequence length L should be adjusted for each repertoire to capture the junction region.
Given a sequence 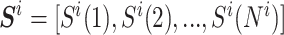 , where Ni is the length of the ith sequence and Si(x) indicates its xth nucleotide, we define the truncated sequence of size L (number of nucleotides) as
, where Ni is the length of the ith sequence and Si(x) indicates its xth nucleotide, we define the truncated sequence of size L (number of nucleotides) as 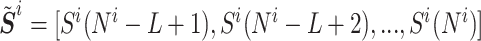 . First, from the 74 artificial datasets, we use a subset of 20 repertoires and seek an optimal value for L, the length of the sequence used for the tf-idf representation, and for k the number of base pairs used for the k-mer representation. The remaining 54 datasets will be used later to evaluate the method’s performance. To compare the performance across different values of L and k, for each value of L in the range [100,190] and k ∈ {2, ..., 9} we first tune the clustering distance threshold to obtain
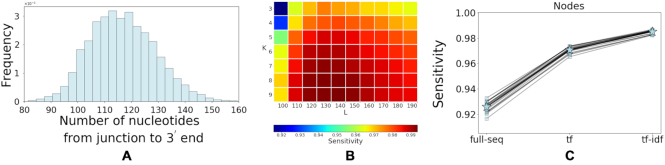
. First, from the 74 artificial datasets, we use a subset of 20 repertoires and seek an optimal value for L, the length of the sequence used for the tf-idf representation, and for k the number of base pairs used for the k-mer representation. The remaining 54 datasets will be used later to evaluate the method’s performance. To compare the performance across different values of L and k, for each value of L in the range [100,190] and k ∈ {2, ..., 9} we first tune the clustering distance threshold to obtain 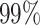 specificity. Then we evaluate the sensitivity and PPV. Based on statistics of aligned sequences from the MS dataset, we expect that this range of lengths will be sufficient to cover all of the J segment along with the adjacent junction region (see Figure 5 (A)). As depicted in Figure 5(B), when L is in the range [120,150] the node sensitivity is peaked. Furthermore, increasing the value of k improves the performance, but also increases memory and computational requirements. The node PPV in this experiment remains higher than 0.99. For the rest of the experiments, we use L = 150 and k = 5 which are sufficient for a node sensitivity >0.98.
specificity. Then we evaluate the sensitivity and PPV. Based on statistics of aligned sequences from the MS dataset, we expect that this range of lengths will be sufficient to cover all of the J segment along with the adjacent junction region (see Figure 5 (A)). As depicted in Figure 5(B), when L is in the range [120,150] the node sensitivity is peaked. Furthermore, increasing the value of k improves the performance, but also increases memory and computational requirements. The node PPV in this experiment remains higher than 0.99. For the rest of the experiments, we use L = 150 and k = 5 which are sufficient for a node sensitivity >0.98.
Figure 5.
(A) 150 nucleotides is sufficient to cover the junction in most sequences. The number of nucleotides from the start of the junction to the 3′ end was calculated for each sequence of 20 artificial repertoires. Note that this count includes additional base pairs from the constant region between the J region and the primer (see Figure 4). (B) Comparing the node sensitivity based on different number of nucleotides (L) and different k-mers values. The threshold is tuned to keep the specificity constant (0.99). The node PPV remains higher than 0.99 across all values of L and k presented above. (C) Performance comparison for three different settings; tf-idf applied to the full sequence (full-seq), tf applied to L=150 nucleotides from each sequence (tf) and tf-idf applied to L=150 nucleotides from each sequence (tf).
Next, we compare the performance of the alignment free method using three different variants of the tf-idf representations. The first term entitled full-seq, in which the tf-idf is applied to the full sequence (with variable length). The second, referred as tf in which only the tf normalization is applied to a fixed part of the sequence with length L = 150. The last, named tf-idf, inputs a fixed part of the sequence as tf does, but uses both the tf and idf normalizations. As in the field of NLP, we speculate that the idf normalization will help by up weighting the unique k-mers, which represent the diverse junction region derived from V(D)J recombination as well as unique SHM that are shared by clonal relatives. At the same time, the idf should down weight the common k-mers, which correspond to the unmutated V, D, and J gene sequences. As in the previous experiment, we use a threshold that achieves 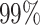 specificity and evaluate the sensitivity of the alignment free method. By applying the tf-idf to full-seq we obtain
specificity and evaluate the sensitivity of the alignment free method. By applying the tf-idf to full-seq we obtain 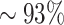 sensitivity, using a fixed part of the sequence improves the performance to
sensitivity, using a fixed part of the sequence improves the performance to 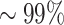 , while without the idf normalization the sensitivity is
, while without the idf normalization the sensitivity is 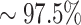 (see Figure 5 (C))
(see Figure 5 (C))
The alignment free has high sensitivity, specificity, and PPV
Next, we compare the overall performance of the alignment free method to the performance of a widely-used junction-based distance method (14) (see Supplementary material for more details) using the remaining 54 simulated repertoires. Comparing to a state-of-the-art-method allows us to evaluate whether the alignment step (along with gene assignment and sub-grouping) is necessary for accurate clonal assignment.
To compare the two methods in terms of sensitivity, specificity, and PPV, we first apply the junction-based distance method (describe in the Methods section) and evaluate performance. For the alignment free method we tune the threshold using the negation approach aiming for a node specificity value of 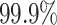 using negation sequences that are randomly selected from (37) as functional BCRs that express either IGHG or IGHM constant regions and do not have any indels. We assign clones based on the estimated threshold and evaluate the alignment free method’s node sensitivity, specificity, and PPV values. The alignment free method achieves ∼
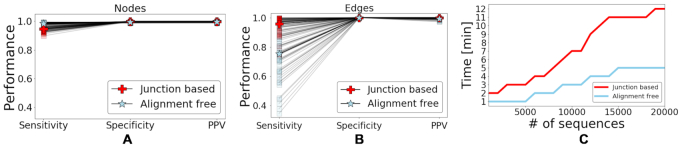
using negation sequences that are randomly selected from (37) as functional BCRs that express either IGHG or IGHM constant regions and do not have any indels. We assign clones based on the estimated threshold and evaluate the alignment free method’s node sensitivity, specificity, and PPV values. The alignment free method achieves ∼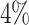 higher node sensitivity compared with the junction-based distance method, while maintaining similar node specificity and PPV (values presented in Figure 6 (A)). However, the junction-based distance method outperforms the alignment free method (see Figure 6 (C)) in terms of edge sensitivity, in part because large clone are sometimes split into smaller groups by the alignment free method.
higher node sensitivity compared with the junction-based distance method, while maintaining similar node specificity and PPV (values presented in Figure 6 (A)). However, the junction-based distance method outperforms the alignment free method (see Figure 6 (C)) in terms of edge sensitivity, in part because large clone are sometimes split into smaller groups by the alignment free method.
Figure 6.
(A) Performance comparison of the alignment free method against the junction-based distance based on 54 artificial datasets. We evaluate each approach by measuring node sensitivity, specificity and PPV. (B) Performance comparison using the same 54 artificial datasets but evaluating edges sensitivity, specificity and PPV. (C) A run time comparison in minutes between the standard junction-based pipeline (red line) and the proposed alignment free procedure (light blue line). The analysis is based on sequences from the simulated repertoires described in the main text (see Section Simulation of Clonal Expansions). The comparison was performed on Yale’s High Performance Computing, using one node. Each node has 20–36 CPU cores with a working frequency of 1.86 GHz. The junction-based procedure involves the application of IgBlast to every sequence followed by the junction-based clonal assignment method (using Change-O-DefineClone function from the SHazaM R package (32)). The alignment free procedure involves cloning first based on the tf-idf representation followed by IgBlast applied to one sequence per clone.
In the Supplementary material we qualitatively demonstrate the efficacy of the negation approach in controlling the false positive rate (specificity). First, in Supplementary Figure S7 we present the distributions of estimated thresholds using negation sequences from two independent studies: (24) and (37). The two distributions overlap, and have means of 0.29 and 0.28 using negations from (24) and (37), respectively. This suggests that the negation approach is not very sensitive to the precise data set used. Next, in Supplementary Figure S7(c) we compare the estimated node specificity to the true node specificity computed based on the simulated repertoires. The results indicate that the node specificity estimated by the negation approach is highly correlated with the true node specificity, but has a slight downward bias.
One advantage of the alignment free method is that it does not require running the full repertoire though a V(D)J assignment program like IgBlast. However, once a clone is identified, it is still important to determine the V, D and J gene assignments for biological interpretation. To achieve this, we randomly select a clone representative that does not contain non-ACGT characters (gaps or N’s) and run it through IgBlast for gene assignment. Thus, gene assignment is performed as a final step and is only applied to a subset of sequences. This can save computational resources as carrying out V(D)J assignment once per clone reduces the number of sequence alignments and gene assignments required. Next, we perform a run-time comparison between the two pipelines. The results (presented in Figure 6 (C)) suggest that for a repertoire of 20,000 sequences a pipeline based on the alignment free method can save approximately half of the computational resources.
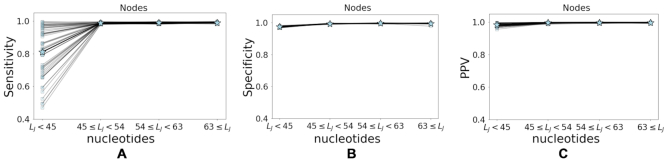
In (14,20) it was observed that the ability to correctly identify clonal relationships drops for BCRs with shorter junction lengths. This drawback was improved in (15) using spectral clustering with an adaptive distance threshold. Here, since we use a single (fixed) threshold to identify clones, we expect that the performance of the alignment free method will deteriorate for sequences with shorter junctions. To evaluate how the length of the junction affects the alignment free method, we apply it to the simulated repertoires, and compute the node sensitivity, specificity and PPV for four different ranges of junction length (LJ), namely (0,45), [45,54), [54,63) and [63,90]. These ranges were selected as they separate the data to approximately equal size subsets. As evident in Figure 7, the alignment free method indeed achieves higher performance when focusing on sequences with longer junctions.
Figure 7.
The alignment free approach performs better for sequences with longer junctions. (A) The node sensitivity, (B) node specificity, and (C) node PPV as a function of the junction length (LJ) evaluation using 54 artificial repertoires. Square marker correspond to performance on each individual dataset, while star markers represent the mean value.
The alignment free method has low false-positive rates on real repertoires
In this section, we compare the alignment free approach to the junction-based distance method on a set of experimentally-derived human repertoires. We use previously published sequencing data from four MS subjects (37) (MS2, MS3, MS4 and MS5). In three of these datasets (MS3-MS5), the automatic distance threshold identification applied by the junction-based distance method (SHazaM-findThreshold (32)) fails, as the distance-to-nearest distribution of these samples is not bimodal. Therefore, following (14) for the junction-based distance method, we use the threshold that was estimated based on MS2 to identify clones in MS3-MS5. The same lack of bimodality appears when evaluating the distance-to-nearest of the tf-idf representation. To estimate the threshold, we use the negation method (explained in the Methods section) with naive sequences from (28). The negation threshold is tuned to obtain 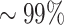 specificity based on MS2. Finally, to have the alignment free method consistent with the setting in (14), we use the same threshold from MS2 for clonal assignments in MS3-MS5.
specificity based on MS2. Finally, to have the alignment free method consistent with the setting in (14), we use the same threshold from MS2 for clonal assignments in MS3-MS5.
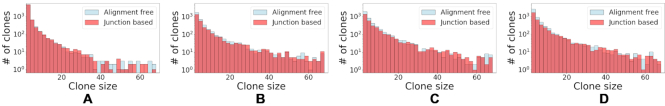
To compare the clonal assignments of the alignment free to the assignments made by the junction-based method, we compute the normalized mutual information (NMI) between the clonal assignment of both methods. The NMI measures how well the two clonal assignments predict one another, and has a highest possible value of 1. As appears in Table 1, the NMI between assignments for all the individuals is high (>0.93), which indicates a high consistency between both clonal assignments methods. As a complementary evaluation, we show that the distribution of clones size is consistent across methods for all four individuals (see Figure 8).
Table 1.
Properties of clones identified by the alignment free method. NMI refers to the Normalized Mutual Information between clonal assignments made by the alignment free method compared to the junction-based distance method. We also determined the percentage of clones with members expression non-unique V or J genes, or non-unique junction lengths
| MS-2 | MS-3 | MS-4 | MS-5 | |
|---|---|---|---|---|
| NMI (with JB) | 0.98 | 0.96 | 0.96 | 0.93 |
| Non unique V [%] | 0.008 | 0.008 | 0.006 | 0.002 |
| Non unique J [%] | 0.038 | 0.036 | 0.054 | 0.014 |
| Non unique Jun. [%] | 0.11 | 0.053 | 0.027 | 0.025 |
Figure 8.
High clustering consistency between the junction-based distance (14) and alignment free methods using sequences from four individuals with MS (8). A comparison between the distributions of clone sizes identified by both methods, (A)–(D) corresponding to MS2–MS5. The y-axis indicates the number of clones identified with a specific size (x-axis).
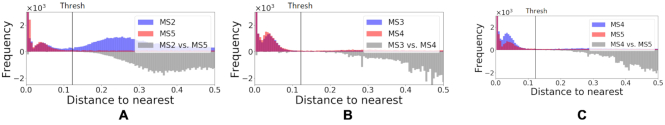
To evaluate the false-positive rates of the alignment free method on real data, we rely on the fact that clones cannot be shared across different individuals. This is because clonally related cells develop from a common ancestor with a single V(D)J germline rearrangement. The false-positive rates are evaluated by first computing the tf-idf based distance-to-nearest sequences across all pairs of individuals in the MS dataset (see Figure 9). Next, we applied the negation approach to automatically estimate the distance threshold (described in the Methods section) by comparing sequences from individuals in the MS data to naive sequences from (28). Here the threshold is tuned to achieve a 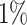 false positive rate based on the negation sequences (i.e. aiming for
false positive rate based on the negation sequences (i.e. aiming for 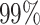 specificity). Clones are defined by cutting the hierarchy based on the estimated threshold, and we count the fraction of clone members from mixed individuals. This portion provides an estimate of the false-positive rates in the MS data. We apply this procedure to all pairs of individuals in the MS data and observe that the false-positive rates are lower than
specificity). Clones are defined by cutting the hierarchy based on the estimated threshold, and we count the fraction of clone members from mixed individuals. This portion provides an estimate of the false-positive rates in the MS data. We apply this procedure to all pairs of individuals in the MS data and observe that the false-positive rates are lower than 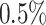 (see Table 2).
(see Table 2).
Figure 9.
Distribution of distance-to-nearest in the tf-idf space. Distances were computed between sequences within one subject (above the x-axis) and between pairs of subjects (below the x-axis), for three pairs of subjects in the MS dataset. The three complement pairs are presented in Supplementary Figure S5.
Table 2.
The alignment free approach was applied to pairs of individuals (MS2-MS4 from (8), rows and columns) and the percentage of sequences predicted to be part of clones that span individuals was calculated. These clonal relationships are considered false positives, as clones cannot be shared across individuals.
| MS-2 | MS-3 | MS-4 | MS-5 | |
|---|---|---|---|---|
| MS-2 | NA | 0.34 [%] | 0.23 [%] | 0.27 [%] |
| MS-3 | 0.34 [%] | NA | 0.032 [%] | 0.012 [%] |
| MS-4 | 0.23 [%] | 0.032 [%] | NA | 0.014 [%] |
| MS-5 | 0.27 [%] | 0.012 [%] | 0.014 [%] | NA |
The alignment free method identifies novel clonal relationships
As the alignment free method is not restricted to a fixed junction or common V or J gene assignments it has the potential to retrieve novel clonal relationships. Here, we evaluate whether the alignment free method identifies such novel relationships in real data. Specifically, we identify clones with multiple V and J gene assignments (when such assignments are made on a sequence-by-sequence basis) or non-unique junction lengths. First we turn our attention toward estimating positive predictive value (PPV) of the alignment free method when used on real repertoires.
Clone members evolve from the same germline; therefore, they should all share the same V and J genes. We use this property to bound the PPV value of the alignment free method. Based on repertoires from the MS study, we observe that the number of clones identified with non-unique J genes is 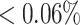 of the total number of identified clones (see the full comparison in Table 1). More significantly, we found that even though we do not use the junction length and only use a small portion of the V gene, the percentage of sequences with non-unique V genes or junction lengths is also low (
of the total number of identified clones (see the full comparison in Table 1). More significantly, we found that even though we do not use the junction length and only use a small portion of the V gene, the percentage of sequences with non-unique V genes or junction lengths is also low (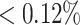 ). If we consider these clonal relationships as errors, they provide an upper bound for the PPV in the MS dataset. However, it is possible that these clonal assignments are correct. For example, sequences with different V or J gene annotations in the same clone could result from incorrect assignments by IgBlast. Such relationships are possible, as the accumulation of SHM can make a BCR derived from single V or J genes seem to stem from distinct V or J genes. Clonal relatives may also have different junction lengths due to the occurrence of indels, which can accumulate as a part of normal SHM (39).
). If we consider these clonal relationships as errors, they provide an upper bound for the PPV in the MS dataset. However, it is possible that these clonal assignments are correct. For example, sequences with different V or J gene annotations in the same clone could result from incorrect assignments by IgBlast. Such relationships are possible, as the accumulation of SHM can make a BCR derived from single V or J genes seem to stem from distinct V or J genes. Clonal relatives may also have different junction lengths due to the occurrence of indels, which can accumulate as a part of normal SHM (39).
To further evaluate whether we have identified true clonal relatives whose initial V or J gene assignments were incorrect or if the non-unique V or J gene assignments are, in fact, false positives, we use a normalized Levenshtein distance (40). The Levenshtein distance (also termed edit distance) finds the minimal number of single edits required to change one sequence to the other. In contrast to the alignment free approach, this comparison takes into account the exact locations in the BCR, and it therefore more accurate than the tf-idf based distance. We note that a direct application of Levenshtein distance to all pairs of sequences is computationally prohibitive. Specifically, for a repertoire of size N, with sequences of length m complexity is O(Nm)2.
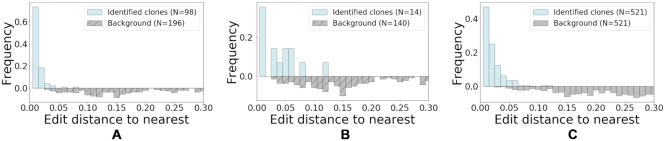
In Figure 10, we first focus on clones with non-unique V or J gene assignments, we present a histogram of a normalized Levenshtein distance (see definition in the Supplementary material and Supplementary Figure S1) between the pairs of closest sequences with different V or J gene assignments. As a background test, we select random groups of sequences, consisting of the same number of sequences as in the retrieved clones. All sequences in such a background group share a V and J gene (majority group) except one sequence with a different gene (minority sequence). Then, we compute the smallest distance between the minority sequence and all sequences in the majority group. The background distribution represents a histogram of such nearest distances (see Supplementary material and Supplementary Figure S2 for more details on this procedure). As evident from Figure 10 (A) and (B), the majority of the clones composed of sequences with non-unique V or J gene assignments identified by the alignment free method have a low distance-to-nearest (relative to the background distribution) when comparing to randomly chosen sequences with different V or J assignments. This supports that a non-negligible portion of the identified sequences are indeed clonally-related, and may have diverged due to SHM. Furthermore, in the Supplementary material, we recompute the distance-to-nearest between the junction part of the sequences (see Supplementary Figure S3), and observe similar low values (relative to the background distribution).
Figure 10.
Evaluation of clones with non unique V gene, J gene or junction length. We use the normalized Levenshtein distance (see definition in the Supplementary material) to asses clones with non-unique V, J, or junction length. (A) Distribution of Levenshtein distance from each sequence to its nearest non-identical neighbor within clones with non-unique J gene assignments (B) or non-unique V gene assignments, or (C) non unique junction lengths. For the background distributions, we generate artificial groups of sequences that share the same V gene, J gene and junction characteristics, and include an additional sequence that differ by one of these characteristics. See supplementary material Supplementary Figure S2 for additional details on this computation.
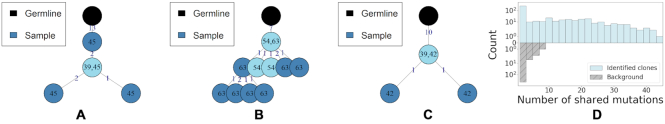
Next, in Figure 10 (C), we show that the distribution of distance-to-nearest among clones with multiple junction lengths is also low (compared to the background distribution), which explains why these sequences were pulled together. A complementary junction-based comparison shows that some of these groups might contain members with highly diverse junctions (see Supplementary material). To further evaluate clones with non unique junction lengths, we study the structure of their phylogenetic trees. Lineage trees were constructed for each clone using Change-O-buildPhylipLineage (version 0.4.5 (32)). The inferred tree topologies (see Figure 11) show that these clones have a non-negligible amount of shared mutations relative to the germline sequence. This is a positive indication that even though these clones have sequences with different junction lengths, they are likely to have evolved from a common mutated ancestor. Furthermore, the average number of minimal shared mutations across the clones with multiple junction length is 7.5 (full distribution appears in Figure 11 (D)). Finally, to corroborate that the sequences presented in Figure 11 have likely evolved from a common ancestor, we present a multi-sequence alignment of these sequences (see Supplementary Figure S4). These results demonstrate the potential of the alignment free method in retrieving clonal relationships between sequences with different junction lengths, or sequences that were assigned to different V or J genes when analyzed individually.
Figure 11.
Tree topologies inferred from clones that include members with multiple junction lengths as determined by the alignment free method (A–C). Branch numbering indicates number of shared mutations, node numbering represents the length of the junction. The germline sequence is colored in black and light blue represents the minority junction length sequences. (D) The distribution of the minimum number shared mutations between sequences with different junction lengths within each clone. The background distribution is computed by sampling random groups of sequences that share the same V and J genes but different junction lengths.
DISCUSSION
B cells play a crucial role in the adaptive immune system. Their ability to recognize and efficiently respond to antigens relies on two diversification mechanisms. The first occurs at an early stage of maturation and acts by joining V and J genes (and D gene for heavy chains) to create a functional antibody receptor. The second part of diversification occurs in the germinal center trough SHM; this step generates a diversified group of clonally related B cells. A critical step in the analysis of high throughput B cell receptor sequencing data is the identification of groups of such clonally related B cells.
We have presented an alignment free method for clonal identification. The approach uses a nucleotide k-mer representation to define a term frequency-inverse document frequency (tf-idf) based distance. This distance is invariant to the exact locations of the k-mers in the BCR sequence; thus, it allows us to bypass the V(D)J alignment step. A second advantage of the alignment free method is that we can identify clonally-related sequences with multiple junction lengths, which can be generated though the accumulation of indels and can be important in affinity maturation to some pathogens. To evaluate the capabilities of this new procedure, we generate simulated repertoires with known clonal relationships between all of the sequences. Using these repertoires, we demonstrate that the alignment free method has high node sensitivity, specificity and PPV. Furthermore, our results suggest that by performing the V(D)J gene assignment after clonal identification, more clone members are retrieved.
We apply the alignment free method to real repertoires collected from four MS subjects. These repertoires lack a correct known clonal assignment; nonetheless, two observed properties suggest a low false positive rate; a low frequency of identified clones containing sequences with different V or J gene assignments, and a low incidence of clones shared across individuals. These features are expected to be enriched among potential false positive relationships. However, it is likely that at least some of them result from incorrect V or J gene assignments by IgBlast, in which cases the alignment free method identifies clone members which would be lost by other approaches.
The library preparation of the MS data is fairly typical; in other experimentally-generated repertoires the precise location of primers may differ and the number of nucleotides required for the tf-idf representation (L) may vary. For more information, see Figure 4, in which we describe the different parts of the BCR and discuss the variability that may arise from different library preparation processes. Our experiments demonstrate that using tf-idf and restricting to a fixed part of the sequence improves node sensitivity, specificity, and PPV values. Nonetheless, the fixed-length used (L) should be adapted to the library preparation technology of each repertoire. Specifically, L should be large enough such that it covers the full junction region (counting nucleotides from the 3′ end). In Figure 5(A) we present the distribution of the number of nucleotides required to cover the junction region based on 20 artificial repertoires.
The final step of the alignment free method requires clustering sequences into clonal groups. Here, we identify these groups by thresholding the dendrogram of distances between sequences. We implement this hierarchical clustering procedure using a fixed threshold, and we optimize this threshold using negation sequences. By computing the distance-to-nearest negation sequences, we optimize a single threshold to obtain high specificity. As demonstrated using simulated repertoires, the performance of the alignment free using a single threshold deteriorates when focusing on sequences with short junctions. A natural extension of this work could alleviate this shortcoming by considering multi-scale thresholds. One example for such solution was presented in (15), where the authors use spectral clustering with an adaptive threshold to identify the clones.
Overall, we have developed an alignment free clonal identification method using tools form natural language processing. We demonstrate using artificial and real repertoires that the alignment free method compares to a state-of-the-art distance-based method, in terms of node sensitivity, specificity and PPV. This shows that the fundamental task of identifying clonal groups does not have to rely on V or J gene assignments. Finally, as the method is capable of identifying clonally-related BCRs with different junction lengths it represents an important improvement in clonal assignments for AIRR-seq analysis.
DATA AVAILABILITY
Source code for this method is freely available at https://bitbucket.org/kleinstein/projects/src/master/Lindenbaum2020/.
Supplementary Material
Contributor Information
Ofir Lindenbaum, Program in Applied Mathematics, Yale University, New Haven, CT, USA.
Nima Nouri, Department of Pathology, Yale School of Medicine, New Haven, CT, USA; Center for Medical Informatics, Yale University, New Haven, CT 06511, USA.
Yuval Kluger, Program in Applied Mathematics, Yale University, New Haven, CT, USA; Department of Pathology, Yale School of Medicine, New Haven, CT, USA; Interdepartmental Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA.
Steven H Kleinstein, Department of Pathology, Yale School of Medicine, New Haven, CT, USA; Interdepartmental Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA; Department of Immunobiology, Yale School of Medicine, New Haven, CT, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01 AI104739 to S.H.K., R01 HG008383 to Y.K., R01 GM131642 to Y.K., R01 GM135928 to Y.K., P50 CA121974 to Y.K., T15LM007056 to N.N.]. Funding for open access charge: NIH [R01 AI104739].
Conflict of interest statement. S.H.K. receives consulting fees from Northrop Grumman.
REFERENCES
- 1. Volpe J.M., Kepler T.B.. Large-scale analysis of human heavy chain V (D) J recombination patterns. Immunome Res. 2008; 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yaari G., Kleinstein S.H.. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med. 2015; 7:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsioris K., Gupta N.T., Ogunniyi A.O., Zimnisky R.M., Qian F., Yao Y., Wang X., Stern J.N., Chari R., Briggs A.W.et al.. Neutralizing antibodies against West Nile virus identified directly from human B cells by single-cell analysis and next generation sequencing. Integ. Biol. 2015; 7:1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosenfeld A.M., Meng W., Chen D.Y., Zhang B., Granot T., Farber D.L., Hershberg U., Luning Prak E.T.. Computational evaluation of B-cell clone sizes in bulk populations. Front. Immunol. 2018; 9:1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meng W., Zhang B., Schwartz G.W., Rosenfeld A.M., Ren D., Thome J.J., Carpenter D.J., Matsuoka N., Lerner H., Friedman A.L.et al.. An atlas of B-cell clonal distribution in the human body. Nat. Biotechnol. 2017; 35:879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenfeld A.M., Meng W., Chen D.Y., Zhang B., Granot T., Farber D.L., Hershberg U., Luning Prak E.T.. Computational evaluation of B-cell clone sizes in bulk populations. Front. Immunol. 2018; 9:1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fukuda T., Chen L., Endo T., Tang L., Lu D., Castro J.E., Widhopf G.F., Rassenti L.Z., Cantwell M.J., Prussak C.E.et al.. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stern J.N., Yaari G., Vander Heiden J.A., Church G., Donahue W.F., Hintzen R.Q., Huttner A.J., Laman J.D., Nagra R.M., Nylander A.et al.. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci. Transl. Med. 2014; 6:248ra107–248ra107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aranburu A., Höök N., Gerasimcik N., Corleis B., Ren W., Camponeschi A., Carlsten H., Grimsholm O., Mårtensson I.-L.. Age-associated B cells expanded in autoimmune mice are memory cells sharing H-CDR3-selected repertoires. Eur. J. Immunol. 2018; 48:509–521. [DOI] [PubMed] [Google Scholar]
- 10. Robinson W.H. Sequencing the functional antibody repertoire—diagnostic and therapeutic discovery. Nat. Rev. Rheumatol. 2015; 11:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoh R.A., Joshi S.A., Liu Y., Wang C., Roskin K.M., Lee J.-Y., Pham T., Looney T.J., Jackson K.J., Dixit V.P.et al.. Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J. Allergy Clin. Immun. 2016; 137:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rubelt F., Busse C.E., Bukhari S.A.C., Bürckert J.-P., Mariotti-Ferrandiz E., Cowell L.G., Watson C.T., Marthandan N., Faison W.J., Hershberg U.et al.. Adaptive Immune Receptor Repertoire Community recommendations for sharing immune-repertoire sequencing data. Nat. Immunol. 2017; 18:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bukhari S. A.C., O’Connor M.J., Martínez-Romero M., Egyedi A.L., Debra Willrett D., Graybeal J., Musen M.A., Rubelt F., Cheung K.H., Kleinstein S.H.. The CAIRR pipeline for submitting standards-compliant B and T cell receptor repertoire sequencing studies to the NCBI. Front. Immunol. 2018; 9:1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta N.T., Adams K.D., Briggs A.W., Timberlake S.C., Vigneault F., Kleinstein S.H.. Hierarchical clustering can identify B cell clones with high confidence in Ig repertoire sequencing data. J. Immunol. 2017; 198:2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nouri N., Kleinstein S.H.. A spectral clustering-based method for identifying clones from high-throughput B cell repertoire sequencing data. Bioinformatics. 2018; 34:i341–i349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Z., Collins A.M., Wang Y., Gaëta B.A.. Clustering-based identification of clonally-related immunoglobulin gene sequence sets. Immunome Research BioMed Central Number 1. 2010; S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glanville J., Kuo T.C., von Büdingen H.-C., Guey L., Berka J., Sundar P.D., Huerta G., Mehta G.R., Oksenberg J.R., Hauser S.L.et al.. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20066–20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kepler T.B. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Research. 2013; 2:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ralph D.K., Matsen IV F.A.. Likelihood-based inference of B cell clonal families. PLoS Comput. Biol. 2016; 12:e1005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nouri N., Kleinstein S.H.. Optimized threshold inference for partitioning of clones from high-throughput B cell repertoire sequencing data. Front. Immunol. 2018; 9:1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nouri N., Kleinstein S.. Somatic hypermutation analysis for improved identification of B cell clonal families from next-generation sequencing data. PLoS Comput. Biol. 2020; 16:e1007977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alamyar E., Giudicelli V., Li S., Duroux P., Lefranc M.-P.. IMGT/HighV-QUEST: the IMGT® web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012; 8:26. [Google Scholar]
- 23. Ye J., Ma N., Madden T.L., Ostell J.M.. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013; 41:W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Briney B., Le K., Zhu J., Burton D.R.. Clonify: unseeded antibody lineage assignment from next-generation sequencing data. Sci. Rep.-UK. 2016; 6:23901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yaari G., Vander Heiden J., Uduman M., Gadala-Maria D., Gupta N., Stern J.N., O’Connor K., Hafler D., Laserson U., Vigneault F.et al.. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front. Immunol. 2013; 4:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang N., He J., Weinstein J.A., Penland L., Sasaki S., He X.-S., Dekker C.L., Zheng N.-Y., Huang M., Sullivan M.et al.. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 2013; 5:171ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hershberg U., Luning Prak E.T.. The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos. Trans. Roy. Soc. B: Biol. Sci. 2015; 370:20140239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vander Heiden J.A., Stathopoulos P., Zhou J.Q., Chen L., Gilbert T.J., Bolen C.R., Barohn R.J., Dimachkie M.M., Ciafaloni E., Broering T.J.et al.. Dysregulation of B cell repertoire formation in myasthenia gravis patients revealed through deep sequencing. J. Immunol. 2017; 198:1460–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou J.Q., Kleinstein S.H.. Cutting edge: ig H chains are sufficient to determine most B cell clonal relationships. J. Immunol. 2019; 203:1687–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smakaj E., Babrak L., Ohlin M., Shugay M., Briney B., Tosoni D., Galli C., Grobelsek V., D’Angelo I., Olson B.et al.. Benchmarking immunoinformatic tools for the analysis of antibody repertoire sequences. Bioinformatics. 2020; 36:1731–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Munshaw S., Kepler T.B.. SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010; 26:867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H.. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015; 31:3356–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramos J.et al.. Using tf-idf to determine word relevance in document queries. Proceedings of the First Instructional Conference on Machine Learning. 2003; 242:Piscataway, NJ: 133–142. [Google Scholar]
- 34. Yun-tao Z., Ling G., Yong-cheng W.. An improved TF-IDF approach for text classification. J. Zhejiang Univ.-Sci. A. 2005; 6:49–55. [Google Scholar]
- 35. Martineau J.C., Finin T.. Delta tfidf: an improved feature space for sentiment analysis. Third International AAAI Conference on Weblogs and Social Media. 2009; [Google Scholar]
- 36. Wang C., Liu Y., Xu L.T., Jackson K.J., Roskin K.M., Pham T.D., Laserson J., Marshall E.L., Seo K., Lee J.-Y.et al.. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J. Immunol. 2014; 192:603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stern J.N., Yaari G., Vander Heiden J.A., Church G., Donahue W.F., Hintzen R.Q., Huttner A.J., Laman J.D., Nagra R.M., Nylander A.et al.. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci. Transl. Med. 2014; 6:248ra107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buckley A.S.C., Mitra M.. Pivoted document length normalization. Proceedings of the 19th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval. 1996; 21–29. [Google Scholar]
- 39. Kepler T.B., Liao H.-X., Alam S.M., Bhaskarabhatla R., Zhang R., Yandava C., Stewart S., Anasti K., Kelsoe G., Parks R.et al.. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe. 2014; 16:304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yujian L., Bo L.. A normalized Levenshtein distance metric. IEEE Trans. Pattern Anal. Mach. Intell. 2007; 29:1091–1095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source code for this method is freely available at https://bitbucket.org/kleinstein/projects/src/master/Lindenbaum2020/.