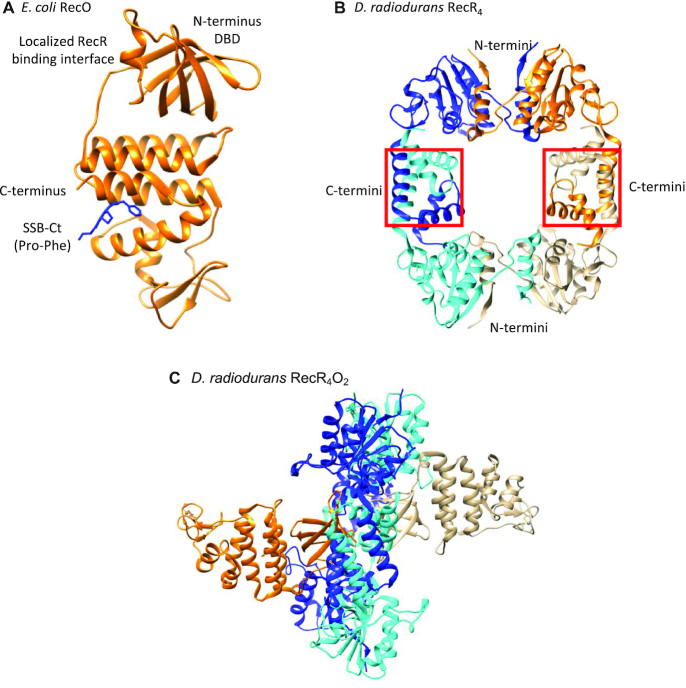
Figure 1.
Structures of RecO and RecR. (A) Crystal structure of E. coli RecO monomer (orange ribbon) bound to a SSB C-terminal acidic tip peptide, WP9 (blue stick) (22,120). Only the last two C-terminal residues of the SSB tip, Pro and Phe, are observed bound to a hydrophobic pocket of the central alpha helical region of RecO. RecO is composed of the N-terminal DNA binding domain, central alpha helical region, and C-terminal zinc binding motif, although zinc is not observed in the E. coli structure. (B) Crystal structure of D. radiodurans RecR tetramer (87). Each RecR monomer is colored in blue, cyan, orange, and gold. D. radiodurans RecR assembles via its C-termini by swapping Walker B motifs and at the N-termini by swapping HhH motifs (87). (C) Crystal structure of ‘closed’ D. radiodurans RecR4O2 complex viewed from the side (90). Two RecO molecules (orange and gold) are bound on each side of the tetrameric ring of RecR (alternating cyan and blue for each subunit). The RecO-RecR interaction site is localized at the N-terminal DBD in RecO (90) and near the central hole in RecR4 ring. Each RecO is situated near the middle of a monomer subunit of a RecR4 ring and interacts with residues from both the N-terminal Walker B motifs and C-terminal HhH motifs, which are important in domain swapping to form a RecR tetramer. RecO is also in contact with residues in the C-terminal Toprim domain.