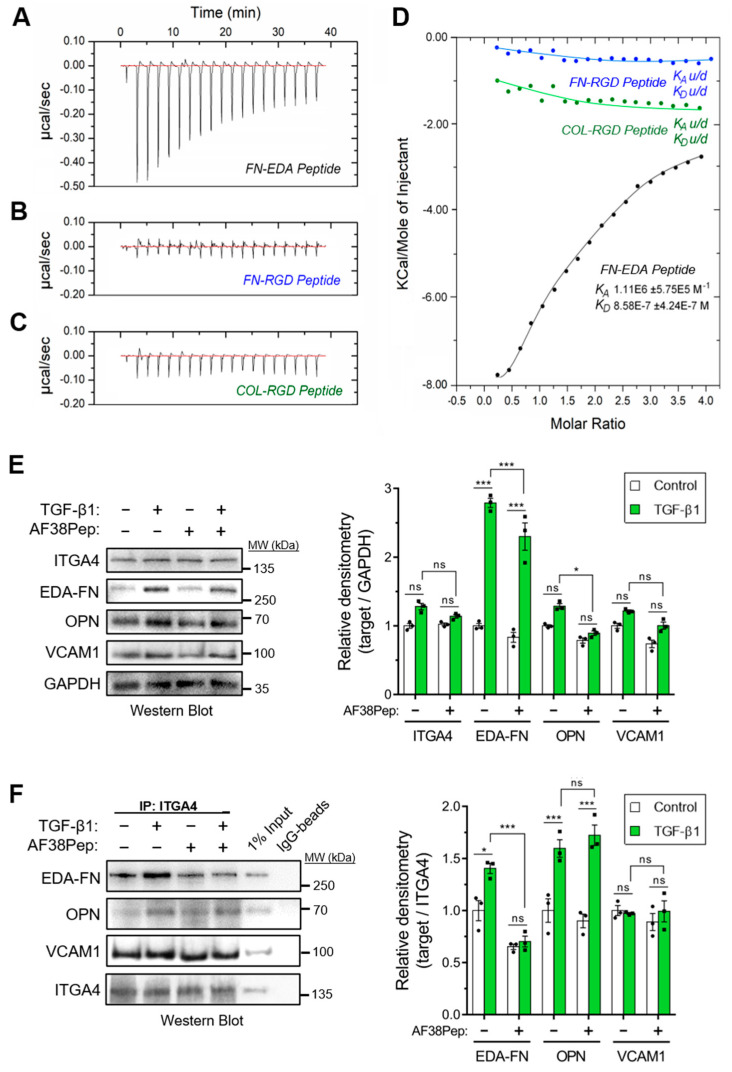
Figure 6.
Binding specificity and blocking function of the synthesized blocking polypeptide. The designed blocking peptide was synthesized alongside 3 small test-binding peptides (FN-EDA, FN-RGD, COL-RGD) and were assessed by isothermal calorimetry (ITC), and calorimetric heat traces were used to test whether the blocking peptide has specificity to bind peptides derived from (A) the predicted EDA binding ligand (FN-EDA, black), (B) the RGD-domain of EDA-FN and FN shown to bind integrin α4β1/β7 at a distal site (FN-RGD, blue), and (C) the RGD-domain of collagen I previously shown to bind different integrin subunits (COL-RGD, negative control, green). (D) The collated isogram is shown alongside predicted dissociation constants (KD) and association constants (KA), calculated using a 1:1 receptor site to ligand site binding mode. (E) Western blot analysis of integrin α4 protein (ITGA4), EDA-FN, osteopontin (OPN), and vascular cell adhesion protein-1 (VCAM1) total protein following 48 h TGF-β1, AF38Pep or TGF-β1+AF38Pep treatments. GAPDH was used as a loading control. Densitometry quantification is shown alongside. (F) Co-IP analysis of ITGA4 binding partners (EDA-FN, OPN, and VCAM1) following 48 h TGF-β1, AF38Pep or TGF-β1+AF38Pep treatments. Data are representative of three independent experiments and displayed as the mean ± S.E. Statistical analysis is shown as *** p ≤ 0.001, * p ≤ 0.05, and ns = no statistical significance (p > 0.05).