Figure 1.
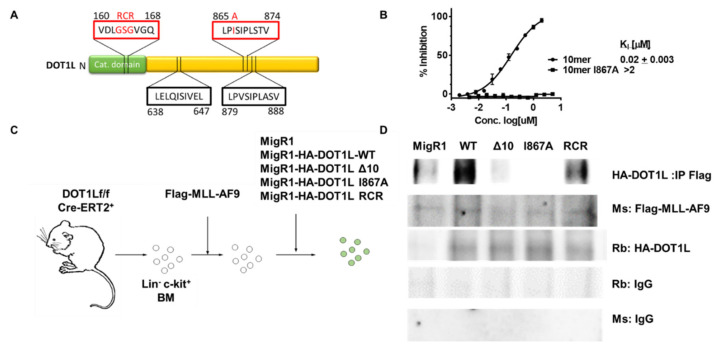
A single amino acid change blocks AF9-DOT1L protein-protein interaction. (A) Protein diagram of DOT1L highlighting the catalytic domain mutation from residues 163–165 and AF9 binding domains at residues 638–647, 865–874 and 879–888. The red boxes indicate regions of the protein that were mutated for the following studies. (B) Fluorescence Polarization assay showing the that the 10mer peptide from residues 865–874 can compete for binding with AF9 with a fluorescein-labeled DOT1L peptide with a KI of 20 nM; whereas, the peptide with the I867A mutation did not show binding up to 10 µM. (C) Schematic of the process of generating DOT1L f/f MLL-AF9 cell lines with different DOT1L constructs reintroduced. Cells were harvested from the bone marrow of DOT1L f/f CreERT2 mice. The bone marrow was lineage depleted and transduced will MLL-AF9. MLL-AF9 were selected by neomycin treatment then subjected to a secondary transduction with either MigR1 as an empty vector control of one of four DOT1L constructs including, DOT1l-WT, DOT1L-Δ10, DOT1L-I867A (Δ10 and I867A mutants block AF9-DOT1L interaction), and DOT1L-RCR (enzymatically inactive DOT1L protein). These cells were then GFP sorted and used for DOT1L excision studies. (D) co-Immunoprecipitation (co-IP) using Flag-MLL-AF9 to pulldown HA-DOT1L in the five established murine cell lines showing disruption of the MLL-AF9 and DOT1L protein-protein interaction in the Δ10 and I867A mutants cell lines as well as the MigR1 cell line where HA-DOT1L is not present with corresponding Rb (rabbit) and Ms (mouse) IgG controls.