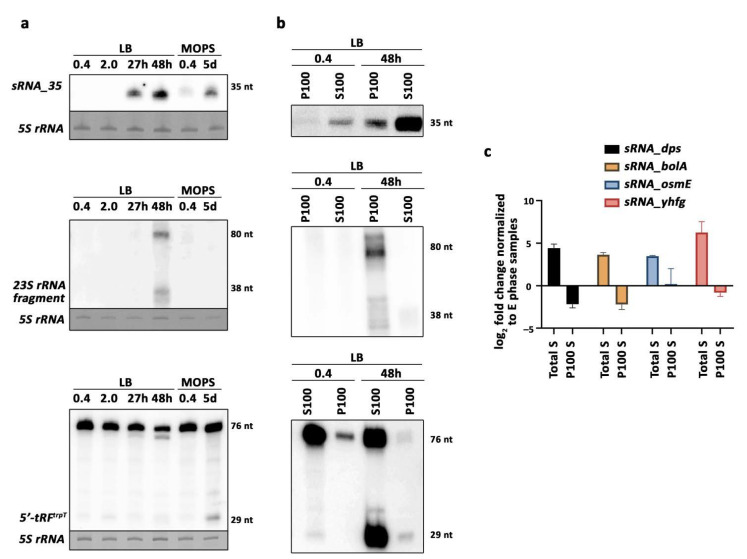
Figure 3.
Validation of identified sRNAs. (a) Northern blot analysis of three candidates: sRNA_35 (FM_+NC_000913.2_4541696_34), 23S rRNA fragment (FM_+NC_000913.2_3942854_40), and 5′-tRFtrpT (FM_+NC_000913.2_3944979_33). 5S rRNA was used as a loading control. Zero point four and 2 were optical density measurements, h = hours, d = days. tRF = tRNA derived RNA fragment. Predicted sizes are displayed on the right in nt (nucleotides). (b) Northern blot analysis of candidates in A. P100 = pellet enriched for ribosome-associated RNA. S100 = supernatant containing non-ribosome associated RNA. (c) RT-qPCR mean and SEM for sRNA_dps (FM_-NC_000913.2_0848114_53), sRNA_bolA (FM_+NC_000913.2_0453657_58), sRNA_osmE (FM_-NC_000913.2_1820262_45), and sRNA_yhfg (FM_-NC_000913.2_3489623_47) in the stationary phase (two biological replicates each). log2 fold change was based on comparison with exponential phase samples for both conditions: Total S = Total RNA samples in the stationary phase. P100 S = ribosome-associated RNA in the stationary phase.