Figure 1.
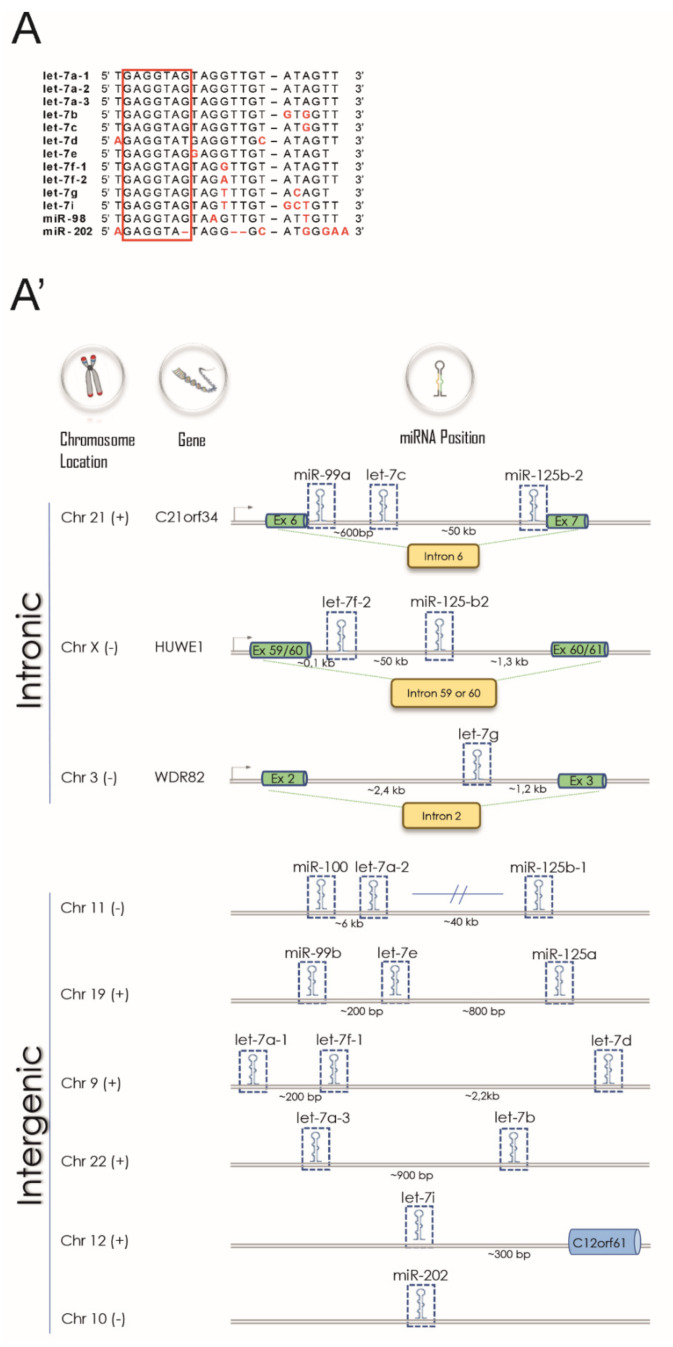
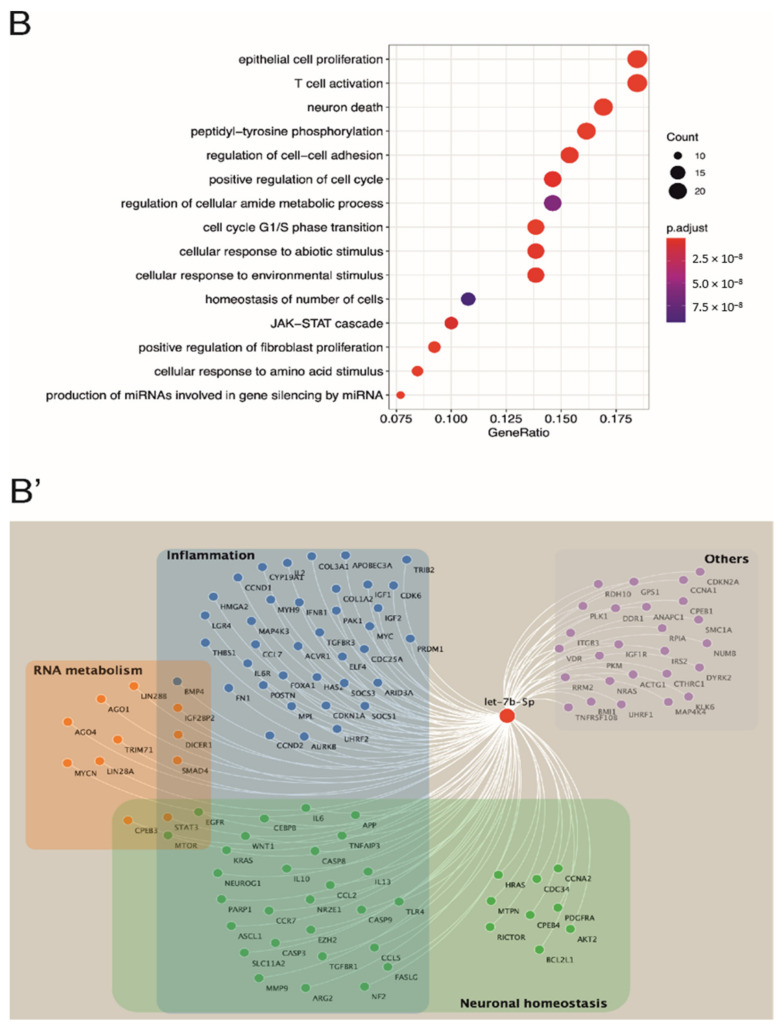
Let-7 family is a good candidate as a MS-associated miRNA. (A) DNA sequences alignment of the mature miRNAs part of the let-7 family. In black the nucleotides positions conserved among all members of the let-7 family. The seed sequence is indicated as a red box. (A’) Genomic organization of let-7 family genes in humans. Gene clusters are divided according to the genome organization (intronic or intergenic). Chromosome strands are indicated by (+) or (−). Figure information sourced from http://microrna.sanger.ac.uk/sequences/. (B) Functional analysis of experimentally validated target mRNAs of let-7 obtained from miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php). The most represented Gene Ontology categories for target mRNAs are reported in the figure. Size dots are correlated with the number of genes that belong to a Gene Ontology category and dots are colored according to the Benjamini-Hochberg false discovery rate adjusted p-values from blue (higher p-adjusted) to red (lower p-adjusted). (B’) Network of let-7 targets that can be ascribed to three main processes involved in MS pathophysiology: inflammation (light blue rectangle); neuronal homeostasis (green rectangle); RNA metabolism (orange rectangle). Target mRNAs of let-7 involved in more than one process are represented into the rectangle overlapping zones. Targets participating in other pathways are grouped into a light violet rectangle (26 out of 130).