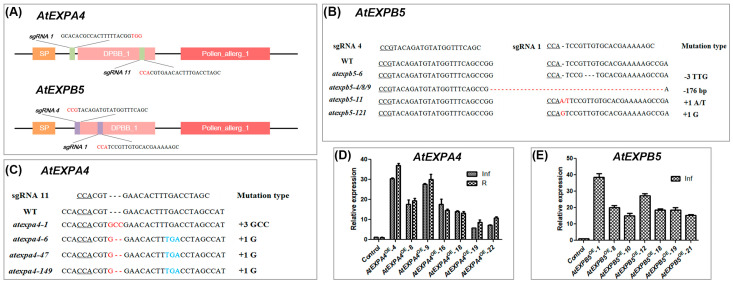
Figure 3.
Confirmation of AtEXPA4 and AtEXPB5 transgenic plants. (A), Genomic location of sgRNAs targeting to AtEXPA4 and AtEXPB5. PAM sites are highlighted in red. (B,C), the gene editing situation of AtEXPA4 and AtEXPB5 by CRISPR/Cas9 system in T3 plants. Underlined letters, red letters, and blue letters represent PAM sites, gene editing sites, and stop codons, respectively. (D), qRT-PCR analysis of AtEXPA4 expression in AtEXPA4OE lines. 35-d-old roots (R) and inflorescences (Inf). BETA-TUBULIN4 was used as the reference gene. AtEXPA4 expression in control lines were normalized to 1. (E), qRT-PCR analysis of AtEXPB5 expression in AtEXPB5OE lines. 35-d-old inflorescences (Inf). BETA-TUBULIN4 was used as the reference gene. AtEXPB5 expression in control lines were normalized to 1. The values are the mean ± SD, three biological replicates with three technical replicates in each biological replicate.