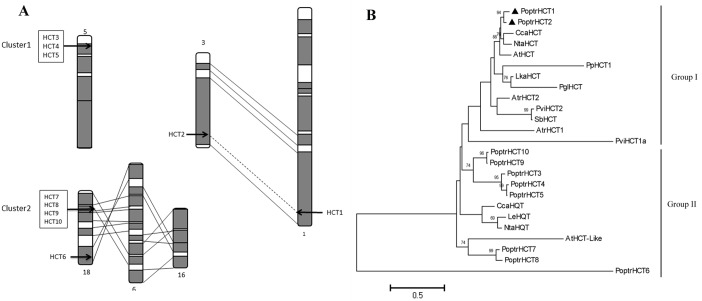
Figure 2. PoptrHCT orthologs organization and phylogenetic analysis.
(A) Organization of HCT orthologs on Populus chromosomes. Regions that are assumed to correspond to homologous genome blocks are shaded gray and connected by lines. The position of genes is indicated with an arrowhead. (B) Phylogenetic analysis of HCT homologs from Populus trichocarpa and other plant species. The PoptrHCT1 and 2 were marked with full black triangle. Two groups for HCT orthologs were shown and HCTs in Group I are likely to transfer hydroxycinnamates to shikimate and have been implicated in monolignol biosynthesis. The scale bar indicates 0.5 amino acid substitutions per site in given length. The accession numbers of sequences used are as followed: Arabidopsis thialiana AtHCT (AT5G48930), AtHCTlike (AT4G29250); Amborella trichopoda AtrHCT1 (ATR_00137G00320), AtrHCT2 (ATR_00727G00010); Cynara cardunculus CcaHCT (DQ104740), CcaHQT(ABK79690); Lycopersicon esculentum LeHQT (AJ582652); Larix kaempferi LkaHCT (AHA44839); Nicotiana tabacum NtaHCT (Q8GSM7), NtaHQT (CAE46932); Picea lauca PglHCT (CZO01061061); Populus trichocarpa PoptrHCT1 (PT01G04290), PoptrHCT2 (PT03G18390), PoptrHCT3 (PT05G02800), PoptrHCT4 (PT05G02810), PoptrHCT5 (PT05G02840), PoptrHCT6 (PT18G03270), PoptrHCT7 (PT18G10470), PoptrHCT8 (PT18G10480), PoptrHCT9 (PT18G10540), PoptrHCT10 (PT18G10550); Physcomitrella patens PpHCT1 (PP00022G00830); Panicum virgatum PviHCT1a (JX845714), PviHCT2 (KC696573 ); Sorghum bicolor SbHCT (XP_002452435.1).