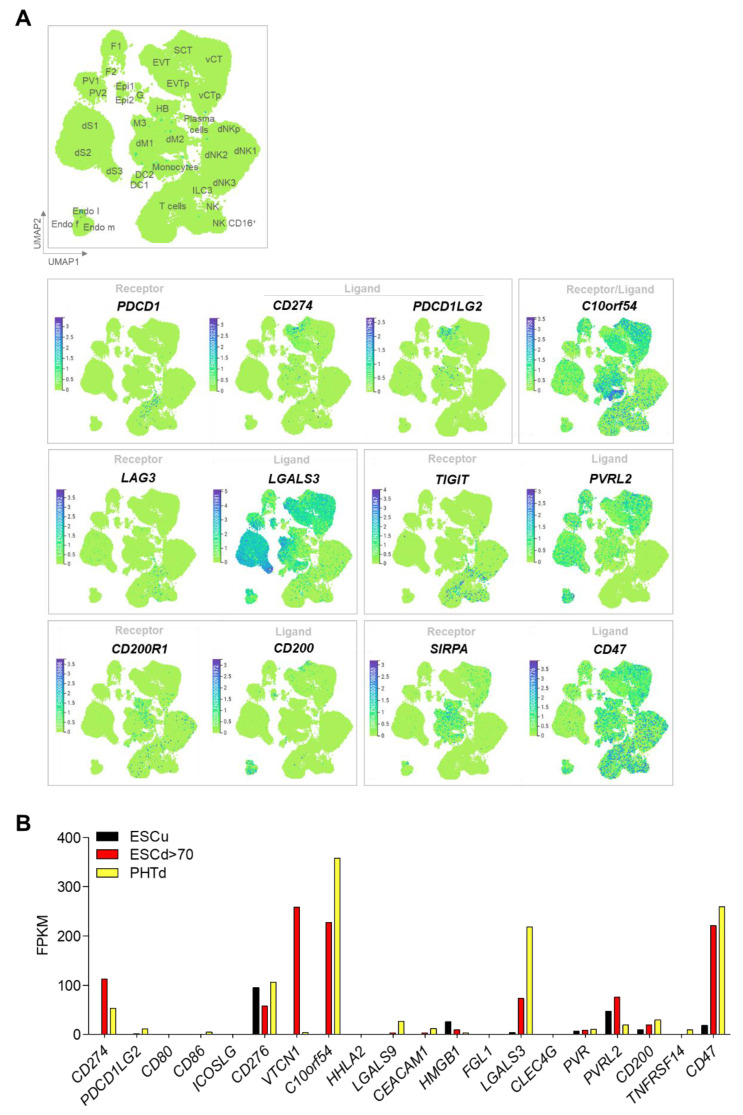
Figure 2.
Expression of inhibitory immune checkpoints. (A) Uniform manifold Approximation and Projection (UMAP) visualization of the log-transformed, normalized expression of immune checkpoint receptors and ligands in placental and decidual cell clusters of the first trimester. Data are available online at http://data.teichlab.org (maternal–fetal interface) (accessed on 11 November 2018) as provided by Vento-Tormo et al. [58]. DC, dendritic cells; dM, decidual macrophages; dS, decidual stromal cells; Endo, endothelial cells; Epi, epithelial glandular cells; F, fibroblasts; HB, Hofbauer cells; PV, perivascular cells; SCT, syncytiotrophoblast; VCT, villous cytotrophoblast; EVT, extravillous trophoblast; f, fetal; ILC, innate lymphocyte cells; l, lymphatic; m, maternal; p, proliferative; M3, maternal macrophages; G, granulocytes. (B) Comparative expression of immune checkpoint inhibitors ligands in undifferentiated H1 ESC (ESCu, black), STB generated from defined DME/F12/KOSR medium that contained BMP4, A83-01, and PD173074 (BAP) treatment-differentiated H1 ESC (ESCd > 70, >70 um size fraction from BAP-differentiated H1 ESC, red), and STB generated from term placenta (PTHd, yellow). Transcript levels are shown in FPKM (Fragments Per Kilobase of transcript per Million mapped reads). RNAseq data are from Yabe et al. [59].