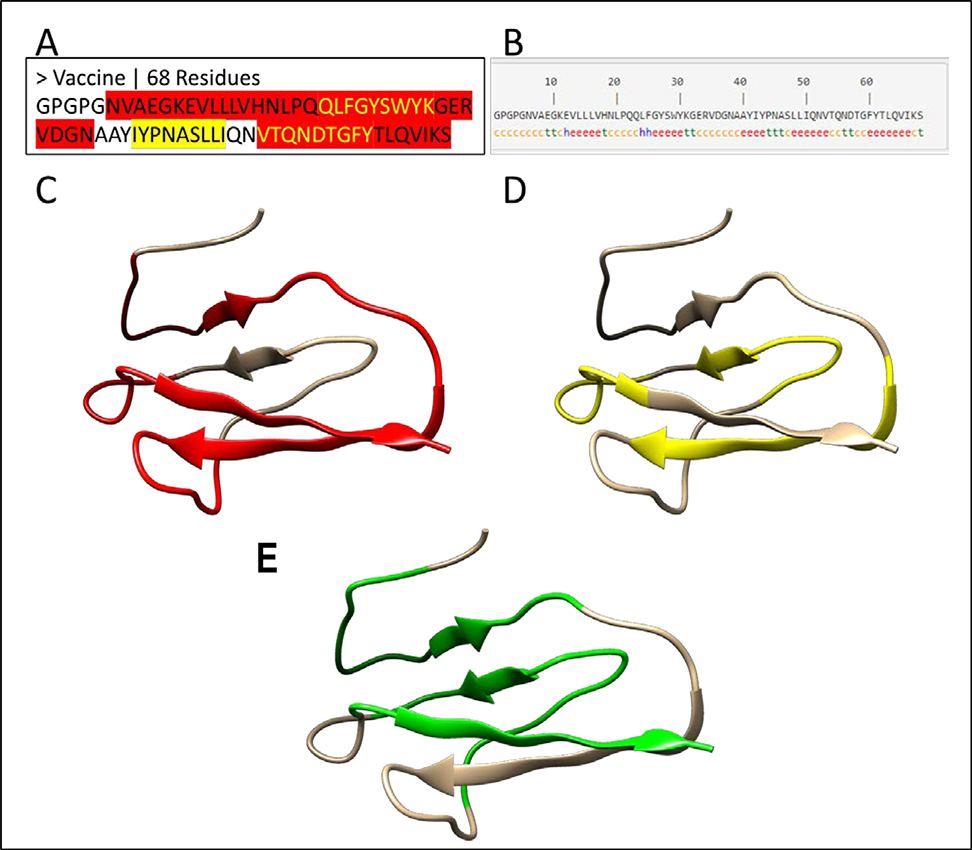
Figure 3: 1-D, 2-D and 3-D structure of a vaccine candidate.
(A) The protein sequence of the vaccine candidate. The MHC I restricted epitopes are highlighted in yellow, while MHC II restricted epitopes are highlighted in red. The yellow text in red regions represents the overlapping epitopes. The MHC I and II epitopes are joined by linker sequences GPGPG and AAY. (B) The second structure of the vaccine candidate. The secondary structure of the vaccine was predicted by the SOPMA server (Geourjon & Deleage, 1995). The letters “h”, “c” and “e” stand for α-helix, random coil and extended β-strand respectively. (C-E) The 3-D structure of the vaccine candidate predicted by a homology modeling method with red color for MHC II epitopes (C), yellow for MHC II epitopes (D), and green for interferon gamma epitopes (E) predicted by IFNepitope server (Dhanda et al., 2013).