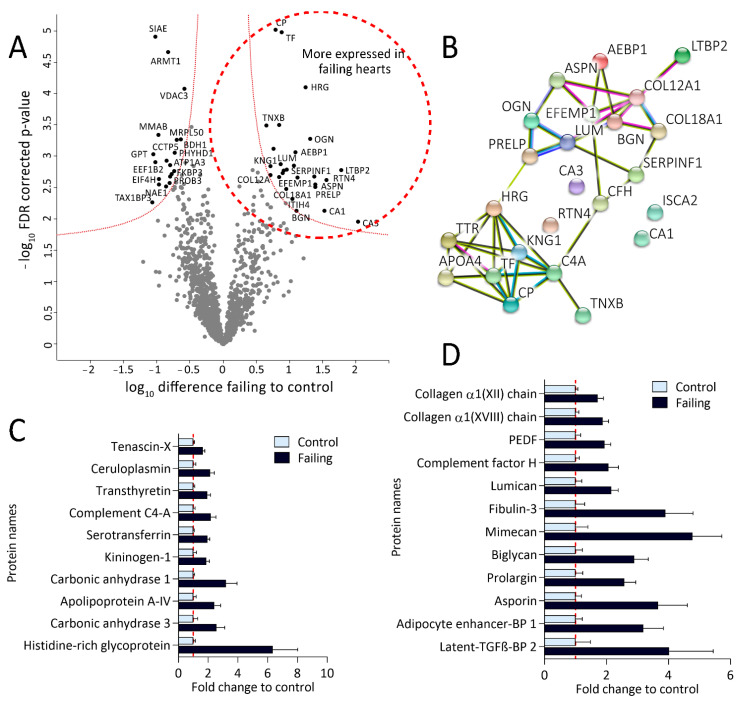
Figure 4.
Proteomic analysis revealed altered protein expression patterns in failing hearts. (A): Volcano plot of proteins identified in failing and control hearts. Significant hits are labeled with their corresponding gene names and depicted in black. Proteins significantly more expressed in failing hearts are shown on the right while the proteins more abundant in the controls are located on the left side of the plot. (B): String protein-interaction analysis of proteins significantly more expressed in failing hearts results in two differential protein clusters (false discovery rate (FDR)-corrected p-value < 0.05, fold change (failing/control hearts) >1.5). (C): Bar plot displaying fold changes of significantly more expressed proteins from the lower String protein cluster (values normalized on the mean of the control group). (D): Bar plot representing fold changes of significantly up-regulated proteins from the upper String protein cluster, namely involved in extracellular matrix remodeling, wound healing, and fibrosis (values normalized on the mean of the control group). PEDF—pigment epithelium-derived factor (gene name: SERPINF1), BP—binding protein.