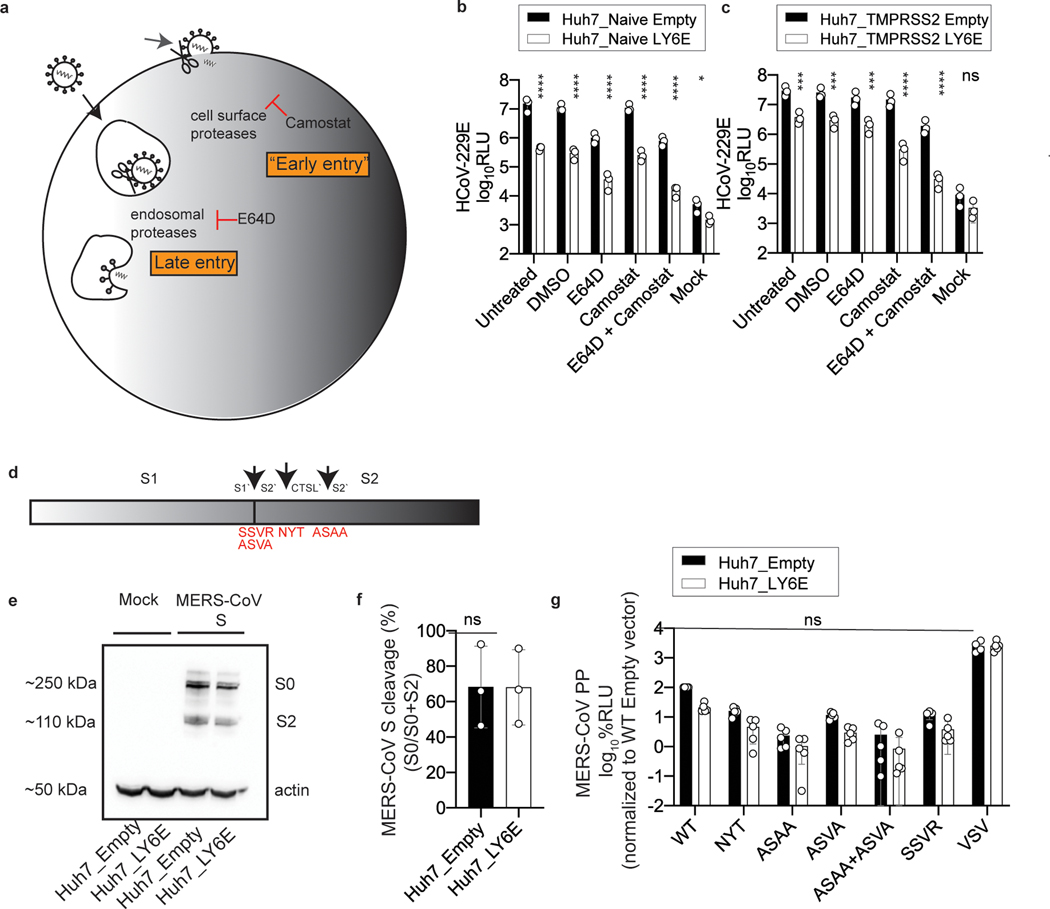
Extended Data Fig. 4. The antiviral effect of LY6E is independent of proteolytic cleavage of CoV spike protein.
a, Schematic depiction of cell entry routes of CoVs and intervention by selected compounds. b-c, LY6E or empty control-expressing Huh7 cells naïve for (b) or ectopically overexpressing TMPRSS2 (c) were pre-treated with the indicated compounds before infection with HCoV-229E-Rluc. d, Schematic depiction of the CoV spike (S) protein containing the subunits S1 and S2. Arrows indicate the S1/S2’ cleavage site, a proposed cleavage site for cathepsin L (CTSL’) and the S2’ cleavage site. Amino acid exchanges disrupting the respective cleavage sites are depicted in red. e, Western blot of S cleavage in LY6E or empty control-expressing Huh7 cells transfected with a plasmid encoding for MERS-CoV S protein (S0= uncleaved, S2= S2 subunit). f, MERS CoV S cleavage was analyzed by quantification of S0 and S2 bands. g, LY6E or empty control-expressing Huh7 cells inoculated with CoV-pseudoparticles (pp) harboring MERS-CoV S WT or MERS-CoV S proteins containing various cleavage site mutations. Data represent mean of independent biological replicates, n=3 (b), n=3 (c), n=3 (e), n=3 (f), n=5 (g). Statistical significance was determined by two-way ANOVA followed by Sidak’s multiple comparisons (b-c, g) or two-tailed unpaired student’s t-test with Welch’s correction (f). Error bars: SD. P values (left-to-right): b, **** p=4.4 × 10−9, p=1.6 × 10−9, p=6.2 × 10−9, p=7.5 × 10−10, p=3.0 × 10−10, * p=0.0179; c, *** p=0.0007, p=0.0005, p=0.0006, **** p=4.8 × 10−8, p=1.8 × 10-8.