Figure EV2. Characterization of the impacts of arginine methylation on METTL14–RNA interactions and RNA methylation activity.
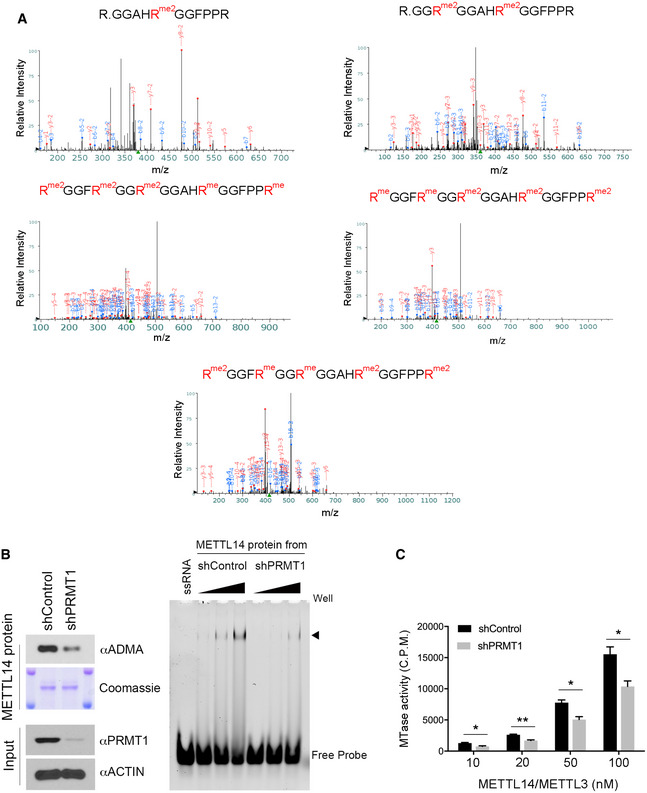
- Identification of METTL14 arginine methylation sites by mass spectrometry. LC‐MS/MS was performed on METTL14 proteins purified from HEK293 cells. Five peptides that are mono‐ or dimethylated were identified (R438, R442, R445, R450, and R456).
- Recombinant METTL14 proteins purified from PRMT1 knockdown HEK293 cells exhibits reduced RNA interactions. Flag‐METTL14 was expressed and purified from control (shControl) and PRMT1 knockdown (shPRMT1) HEK293 cells. The methylation level of METTL14 was detected by Western blot using an anti‐ADMA antibody. The amount of protein was visualized by Coomassie staining (left panel). EMSA was performed to compare the interaction of recombinant METTL14 purified from shControl and shPRMT1 HEK293 cells with 6‐FAM‐labeled RNA. Arrow indicates the shift of the RNA probe caused by the protein–RNA interaction (right panel).
- Recombinant METTL14 purified from PRMT1 knockdown HEK293 cells exhibits reduced RNA methylation activity. In vitro RNA methylation assays were performed by incubating biotin‐labeled RNA substrates with METTL3/METTL14 methyltransferase complexes purified from control (shControl) and PRMT1 knockdown (shPRMT1) HEK293 cells. The enzymatic activity was measured in counts per minute (c.p.m.) using a scintillation counter. Data from three independent replicates were analyzed by Student’s t‐test and shown as mean ± SD. *P < 0.05; **P < 0.01.
Source data are available online for this figure.
