Figure EV3. Localization and interaction analysis of arginine methylation‐deficient METTL14 in cells.
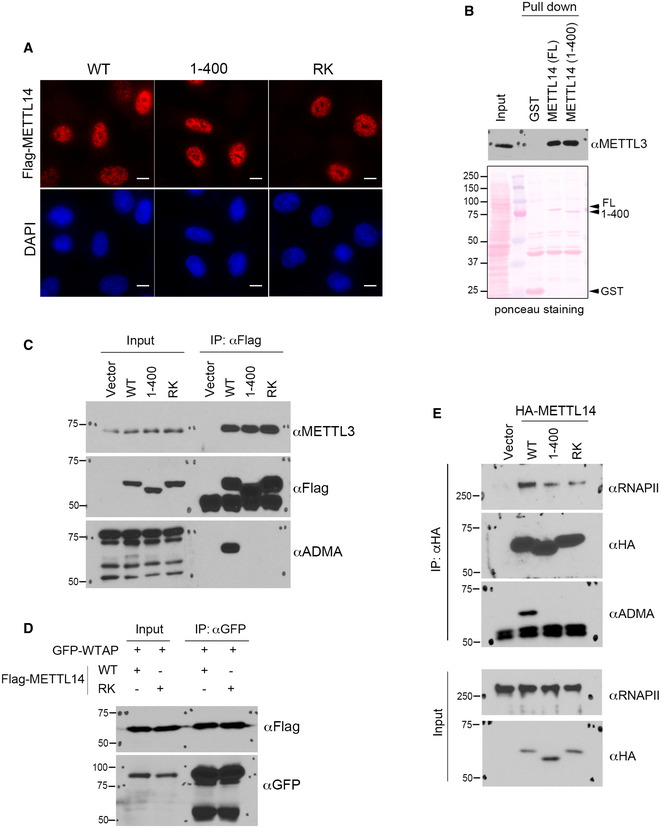
- Immunofluorescence assays were performed to examine the subcellular localization of WT, C‐terminal IDR‐truncated (1–400), and arginine methylation‐deficient (RK) mutant METTL14 in HeLa cells. The localizations of all three proteins were detected by using an anti‐Flag antibody. DAPI staining was performed to mark the cell nucleus. Scale bar: 10 µm.
- GST pull‐down assays were performed by incubating GST‐tagged full‐length (FL) and C‐terminal IDR‐truncated (1–400) METTL14 with HEK293 cell lysate. Western blot analysis was performed using an anti‐METTL3 antibody. Ponceau S staining shows the loading of the recombinant proteins in the pull‐down samples.
- Co‐IP assays were performed to detect the interactions of WT, C‐terminal IDR‐truncated (1–400), and arginine methylation‐deficient (RK) mutant METTL14 with METTL3. The methylation of METTL14 protein was confirmed by Western blot analysis using an anti‐ADMA antibody.
- Co‐IP assays were performed to detect the interactions of WT and arginine methylation‐deficient (RK) mutant METTL14 with GFP‐WTAP. The HEK293 cells were transiently transfected with the indicated plasmids 48 h before the assays were performed.
- Co‐IP assays were performed to detect the interactions of WT, C‐terminal IDR‐truncated (1–400), and arginine methylation‐deficient (RK) mutant METTL14 with RNAPII. The methylation of METTL14 protein was confirmed by Western blot analysis using an anti‐ADMA antibody.
Source data are available online for this figure.
