Figure EV5. METTL14 C‐terminal IDR arginine methylation regulates m6A deposition on DNA repair genes.
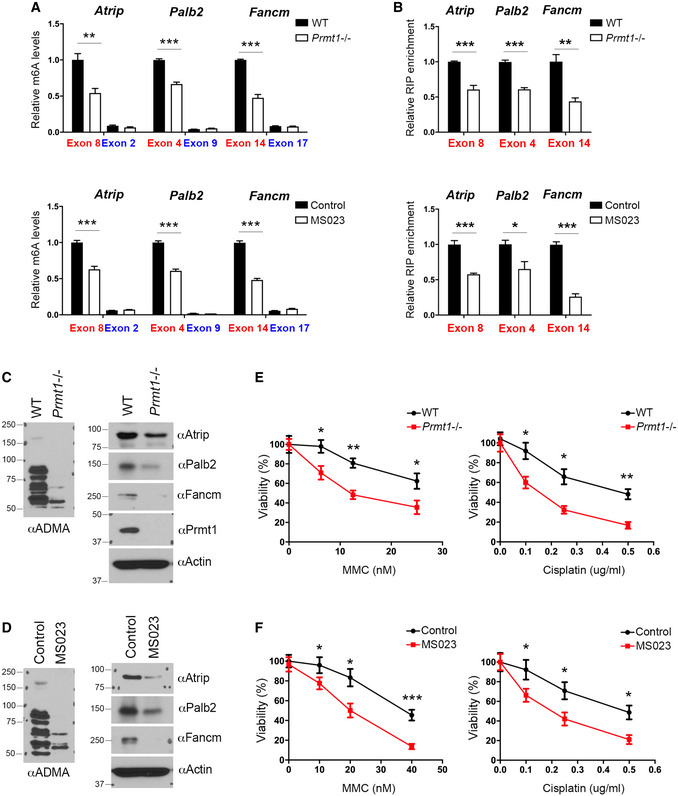
- MeRIP (m6A‐IP)‐qPCR assays were performed for WT and Prmt1 KO mESCs (upper panel), as well as for control and MS023‐treated mESCs (lower panel), to detect the impact of PRMT1 loss or inhibition on m6A deposition at targeted transcripts. m6A‐negative regions of the transcripts (blue) were included as negative controls.
- METTL14 RIP‐qPCR assays were performed for WT and Prmt1 KO mESCs (upper panel), as well as for control and MS023‐treated mESCs (lower panel), to detect the impact of PRMT1 loss or inhibition on the interactions of METTL14 with targeted transcripts.
- The expression of ICL repair genes is reduced in Prmt1 KO mESCs. Total cell lysates from WT and Prmt1 KO mESCs were subjected to Western blot analysis using the indicated antibodies.
- The expression of ICL repair genes is reduced in MS023‐treated mESCs. Total cell lysates from control and MS023‐treated mESCs were subjected to Western blot analysis using the indicated antibodies.
- Knockout of Prmt1 sensitizes mESCs to ICL damage. WT and Prmt1 KO mESCs were treated with various concentrations of MMC (left panel) or cisplatin (right panel) for 4 days before cell viability was measured.
- Similar to (E), except that mESCs were treated with MS023, to inhibit type I PRMT activity, while they were treated with MMC (left panel) or cisplatin (right panel).
Data information: In (A), (B), (E), and (F), data from three independent replicates were analyzed by Student’s t‐test and shown as mean ± SD. In both (E) and (F), each point represents the average of three independent replicates and error bars represent standard deviation. *P < 0.05; **P < 0.01, ***P < 0.001.
Source data are available online for this figure.
