Figure 3. The CM upstream region of CENP‐C stabilizes the CENP‐A nucleosome binding.
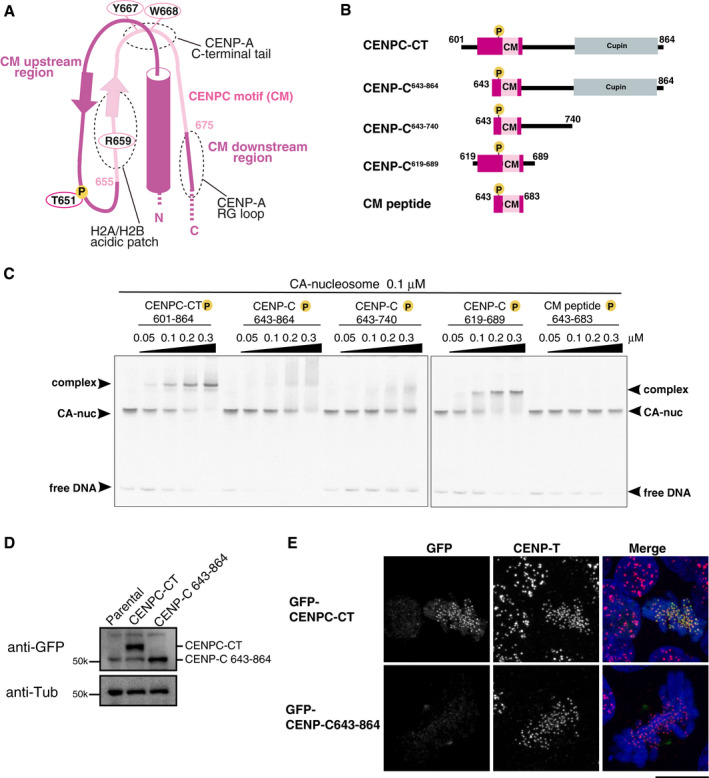
- Schematic representation of the structure of the CENP‐A nucleosome‐binding region in CENPC‐CT. The canonical CENPC motif segment is shown in pink. The CDK1 phosphorylation site (T651) is also indicated. The CM upstream and downstream regions are shown in magenta. Dot circles indicate the recognition sites for H2A/B acidic patch, CENP‐A C‐terminal tail, and RG loopCENP‐A.
- Schematic diagrams of deletion mutants of chicken CENPC‐CT. The fragments contain the canonical CENPC motif and the CM downstream region. CENP‐C643‐864, CENP‐C643‐740, and CM peptide lack the CM upstream region. CENPC‐CT and CENP‐C619‐689 contain the CM upstream region.
- EMSA results of the binding affinities of CENP‐C fragments indicated in (B) to the CENP‐A nucleosome. All fragments were phosphorylated by CDK1.
- Stable expression of GFP‐fused CENPC‐CT and CENP‐C643‐864 (shown in (B)) in CENP‐C knockout chicken DT40 cells. α‐Tubulin (Tub) was probed as a loading control.
- Localization analysis of GFP‐fused CENPC‐CT and CENP‐C643‐864 lacking the CM upstream region (green) on mitotic chromosomes in CENP‐C knockout DT40 cells. CENP‐T (red) was used as a centromere marker. DNA was stained using DAPI (blue). Scale bar indicates 10 μm.
