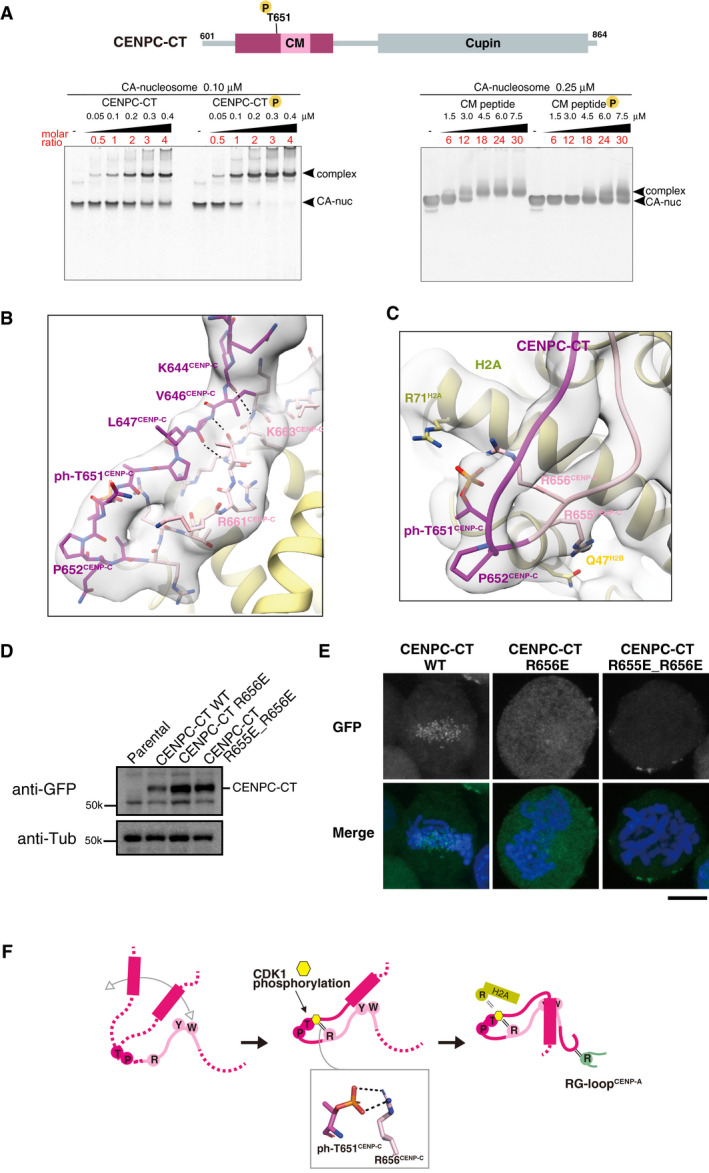
The position of a CDK1 phosphorylation site, T651CENP‐C, is indicated in the diagram of chicken CENPC‐CT. EMSA was performed to examine the binding affinities of phosphorylated or nonphosphorylated CENPC‐CT (left panel) and CM peptide (right panel) to the CENP‐A nucleosome.
Structure of the loop region harboring phosphorylated T651CENP‐C. CENPC‐CT is shown in magenta, except for the pink canonical CENPC motif. The cryo‐EM density map is overlaid on the cartoon model. The main chain interactions forming a short β‐sheet are indicated by black dotted lines.
Magnified view of the structure around phosphorylated T651CENP‐C. Key residues in this region are shown as stick models. The side chains of R656CENP‐C and R71H2A are situated in the vicinity of the phosphoryl group of T651CENP‐C in the distance ranges of 3.5‐4 Å and 3.5‐4.5 Å, respectively.
All constructs analyzed in (E) were stably expressed in CENP‐C knockout cells.
Localization analysis of GFP‐fused CENPC‐CT, CENPC‐CTR656E, and CENPC‐CTR655E_R656E (green) on mitotic chromosomes in chicken DT40 cells. DNA was stained using DAPI (blue). Scale bar indicates 10 μm.
Model indicating the functional role of T651CENP‐C phosphorylation in the CENP‐A‐CENP‐C interaction. Primarily, the CENPC motif associates with CENP‐A nucleosome. Phosphorylation of T651CENP‐C by CDK1 facilitates the loop conformation in the CM upstream region through an intramolecular bridge with the side chain of R656CENP‐C. Subsequently, the N‐terminal α‐helix is folded over to stabilize the CENP‐C‐CENP‐A nucleosome interfaces mediated by the CENPC motif and CM downstream regions.