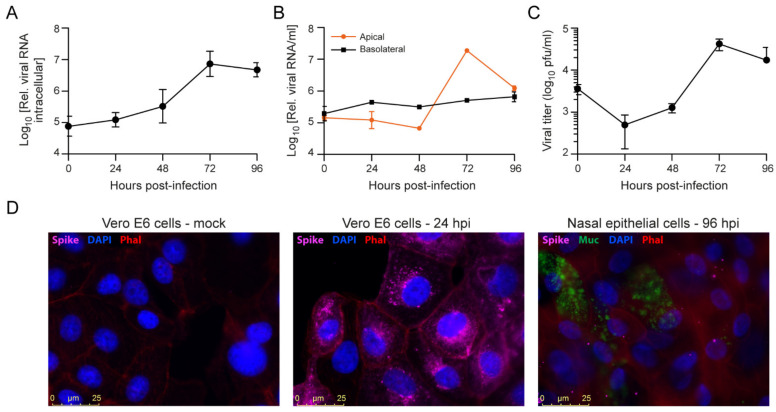
Figure 4.
SARS-CoV-2 infection of primary nasal epithelial cells. Primary nasal epithelial cells, cultured on an air-liquid interface, were infected with SARS-CoV-2 (BavPat1 isolate) at an MOI of 10. At the indicated time points, cells were harvested and viral RNA from (A) cells or (B) apical and basolateral compartment supernatants was quantified by RT-qPCR. Intracellular viral RNA levels were normalized to the human β-actin housekeeping gene. Error bars indicate SD (n = 2). (C) Infectious viral titers from apical supernatants corresponding to the indicated time points were quantified by plaque assay. Error bars indicate SD (n = 2). (D) Immunofluorescence staining of mock (left panel), SARS-CoV-2 infected Vero E6 cells fixed at 24 hpi (MOI = 0.01; middle panel) or SARS-CoV-2 infected primary nasal epithelial cells fixed at 96 hpi (MOI = 10; right panel). The presence of SARS-CoV-2 Spike protein subunit S1 (pink) was assessed in cells stained with DAPI (nucleus, blue) and phalloidin (F-actin, red). Nasal epithelial cells were additionally stained with anti-Muc5AC antibodies (Muc, goblet cells, green). Bar represents 25 µm.