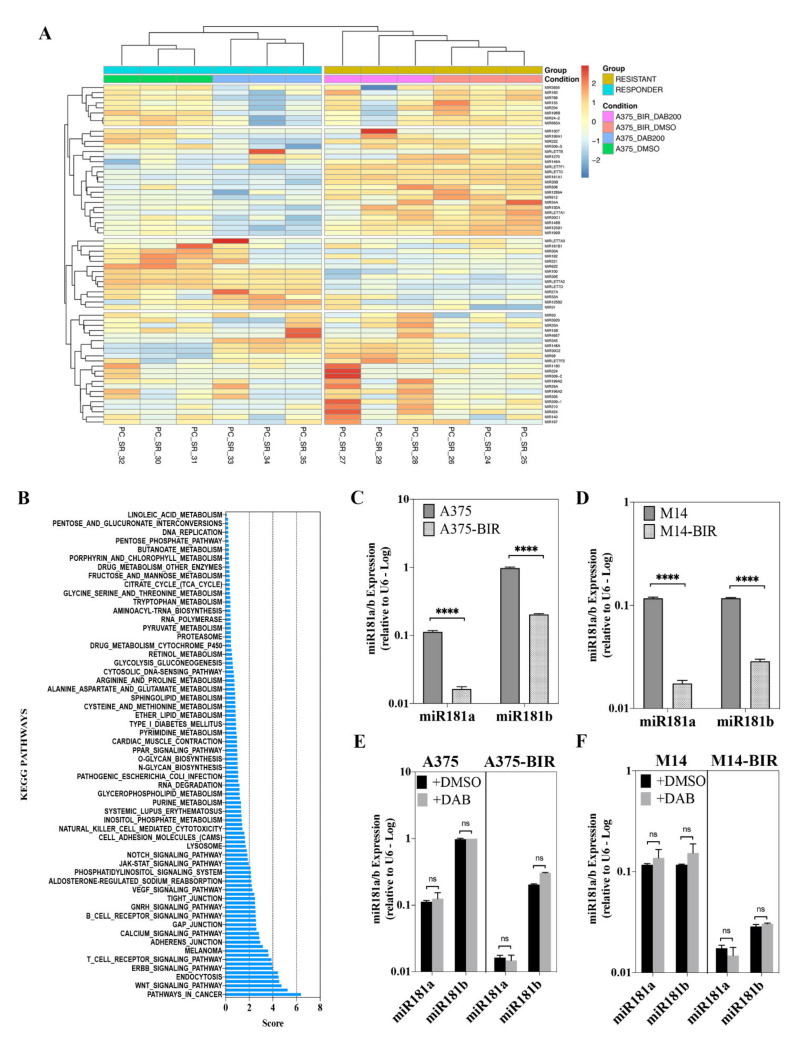
Figure 1.
(A) Hierarchical clustering analysis and heat map based on the expression profiles of 65 miRNAs in responding and non-responding melanoma cells to dabrafenib treatment. Cluster analysis grouped samples and miRNAs according to similarity in expression. miRNAs are in rows and samples in columns. The miRNA clustering tree is indicated on the left and the sample clustering tree is at the top. Red color represents up-regulated expression and blue marks downregulated genes. Yellow indicates resistant cells and blue indicates responsive cells. miRNA, microRNA. To create the heatmap we converted the read counts into log2-counts-per-million (logCPM) values. (B) Putative target genes of differentially expressed miRNAs were obtained from TargetScan and used for KEGG pathway enrichment analysis. Only pathways with an adjusted p value < 0.01 were considered and listed according to a decreasing value of the combined score. (C,D) The expression levels of miR-181a and b in A375, A375-BIR, M14 and M14-BIR melanoma cells, measured by quantitative Real-Time PCR (qRT-PCR). (E,F) The expression levels of miR-181a and b in A375, A375-BIR, M14 and M14-BIR melanoma cells, treated with 200 nM dabrafenib (DAB) and vehicle control (DMSO), measured by qRT-PCR. The miRNA expression levels were normalized to the internal control U6. **** p < 0.0001; ns, not significantly.