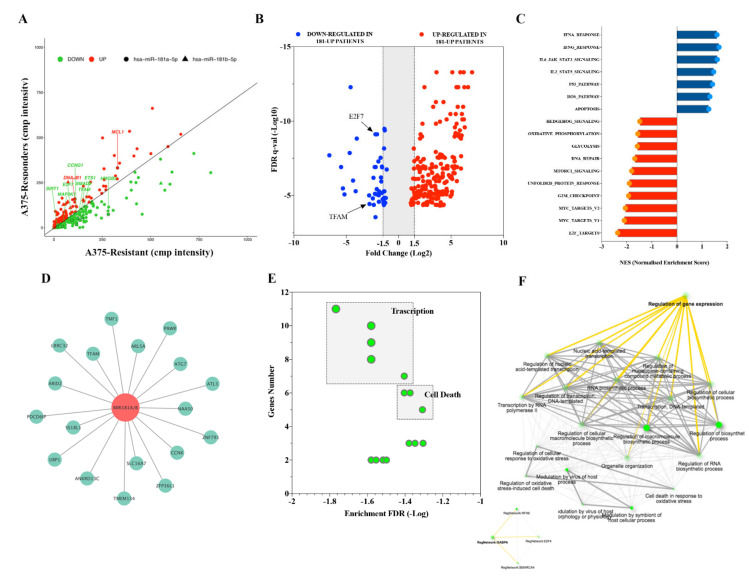
Figure 5.
(A) Scatter plot of the RNA-Seq expression data showing differentially expressed miR-181a and -181b-target genes (DEG) between A375 (responders) and A375-BIR (resistant) cell lines. Dots represent DEGs that are: significantly overexpressed (red) and down-regulated (green). The most critical upregulated and downregulated DEGs targets are indicated. (B) Volcano plot showing the DEGs up (red dots) and down-regulated (blue dots) in patients expressing high levels of miR-181a and -181b (responders group). FDR: False discovery rate. (C) Plot of functional gene set enrichment analysis (GSEA) indicating hallmark gene sets significantly modulated in melanoma patients expressing high levels of miR-181a and -181b (responders group). (D) Network of miRNA-181a- and -181b-negatively regulated predicted target genes in melanoma patients (responders group) visualized by Cytoscape. Green dots represent predicted target genes and the red dot represents hub miRNA. (E,F) GO term enrichment analysis of miRNA-181a- and -181b-negatively regulated predicted target genes in melanoma patients (responders group). (E) Scatter plot of enriched GO pathways according to numbers of genes and FDR. (F) GO network analysis plot shows the relationship between enriched pathways. Darker green nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Most critical networks are highlighted in yellow.