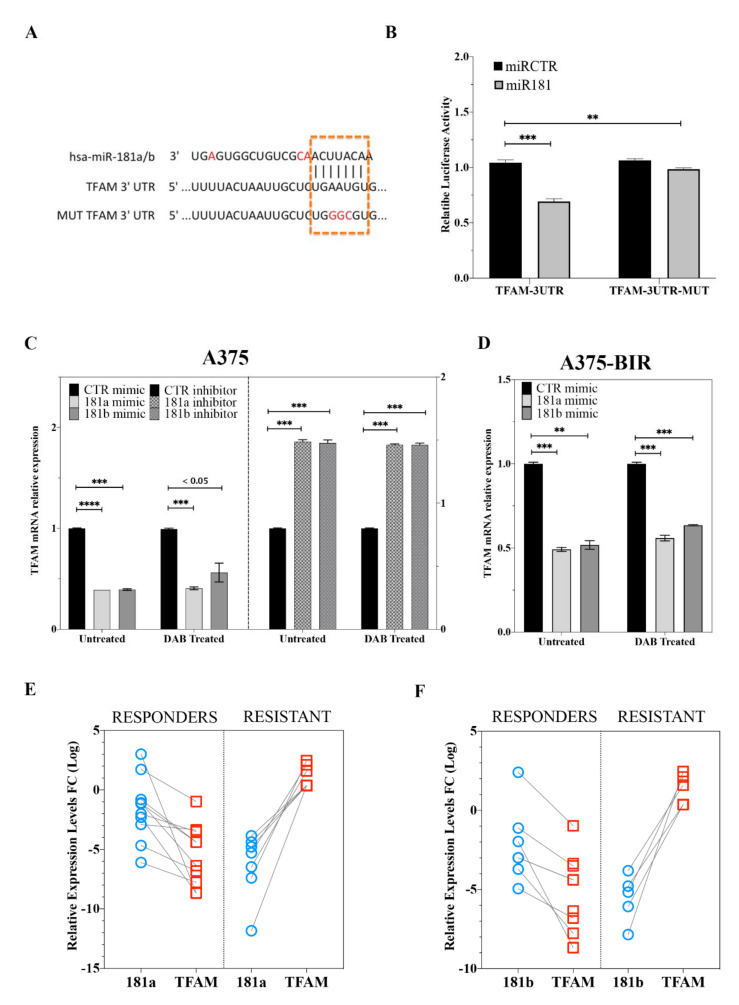
Figure 6.
(A) TFAM 3′UTR contains miR-181a and -181b binding sites. Red-labelled nucleotide indicate the differences between the sequences of miR-181a and -181b. (B) The dual luciferase reporter assay was performed with HeLa cells as described in Materials and Methods. Briefly, HeLa cells were transfected with miR-181a or miR mimic negative control together with the plasmid encoding for the Firefly luciferase (FFL) carrying part of the TFAM 3′-UTR in its wild type form (TFAM-3UTR) or TFAM 3ʹ-UTR with the mutagenized form of the miR-181a and -181b binding site (TFAM-3UTR-MUT). FFL activities were internally normalized to Renilla luciferase activities yielding relative light units (RLU). Histograms show the mean +/− SE from 3 to 6 independent experiments upon normalization to the miR mimic control. ** p < 0.01; *** p < 0.001 in un-paired t-test. (C,D) TFAM mRNA expression analysis in A375 (C) and A375-BIR (D) cells transfected with miR-181a, -181b mimics or their inhibitors or relative controls. Histograms show the mean +/− SE from 3 independent experiments upon normalization to the miR mimic control. ** p < 0.01; *** p < 0.001; **** p < 0.0001 in un-paired t-test. (E,F) Treatment resistant and responder melanomas display miR-181a/b/TFAM axis deregulation. Lower miR-181a (E) and -181b (F) and higher TFAM expression levels in patients resistant during therapy compared to tumors from the responder group. p < 0.001 by Mann-Whitney U test.