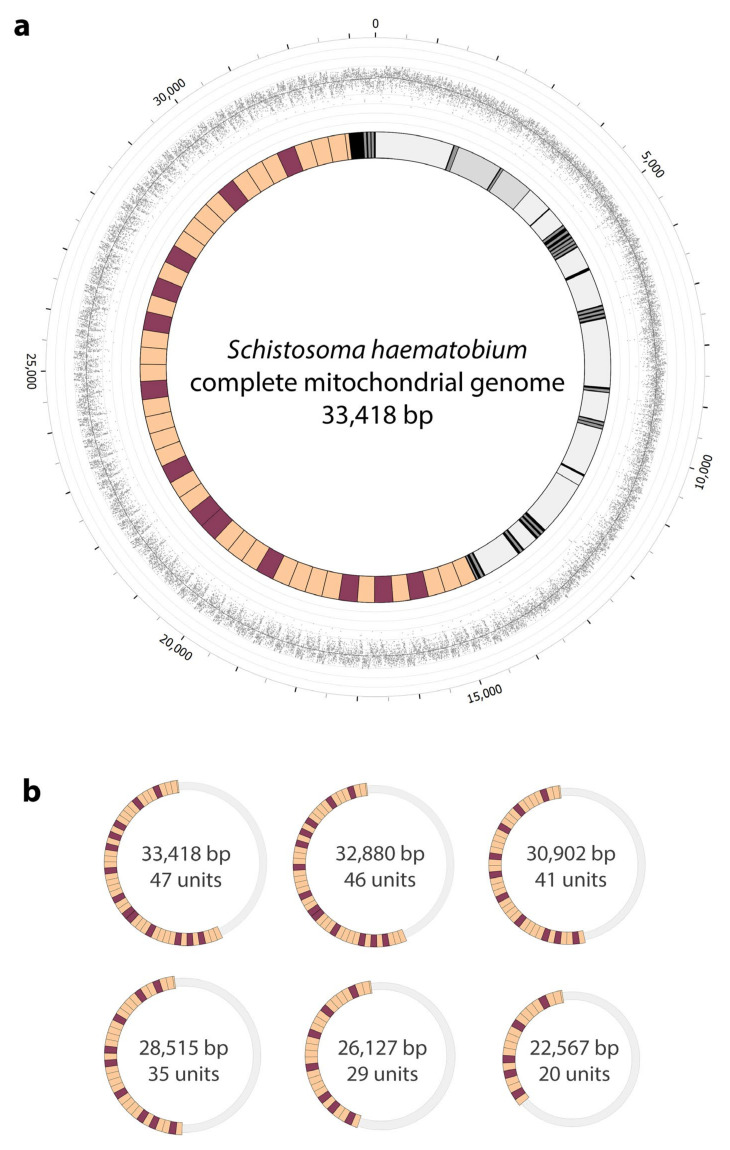
Figure 2.
Complete mitochondrial (mt) genomes representing Schistosoma haematobium. (a) Schematic representation of the dominant, representative mt genome of S. haematobium (33,418 bp; inner circle), including the newly-identified tandem-repeat region (18,458 bp; units ShR1 and ShR2 in orange and purple, respectively). The 12 protein-coding genes, 2 rRNAs and 22 tRNAs (in shades of grey) are in accord with a published reference mt genome available in GenBank (accession no. DQ157222; [22]); short non-coding regions (<350 bp) are in black. The outer circle represents the coverage of long-reads (produced by Oxford Nanopore sequencing) across the genome. The graph shows the depth of nucleotides at each position (grey dots) and the smoothed average of depth across the genome (solid dark grey line). Circular axes represent every 100 reads mapped. Numbers on the outer circle represent positions on the genome in base pairs. (b) Schematic representation of all established mt genome lengths in S. haematobium supported by >150 Nanopore long-reads (cf. Figure 1). Numbers inside represent the length of the mt genome and the number of units in the tandem-repeat region. Sizes of the circles are proportional to genome lengths.