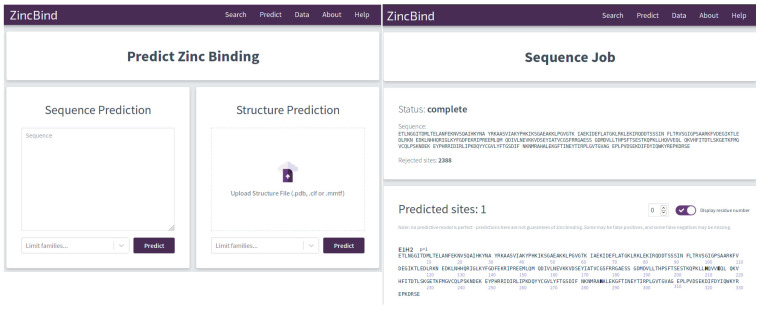
Figure 1.
The graphical interface for the predictors is shown on the left. The user can enter a protein sequence, or upload a structure file. In both cases, the user has the option of limiting the zinc binding families for which the predictor will search, which can save a considerable amount of time. Results of the prediction are shown on the right with the residues predicted to form a binding site shown in bold. This interface consumes the ZincBindPredict GraphQL application programming interface (API), which is also publicly available.