Figure 2.
The synonymous BAP1 mutation do not affect protein stability at the cDNA level
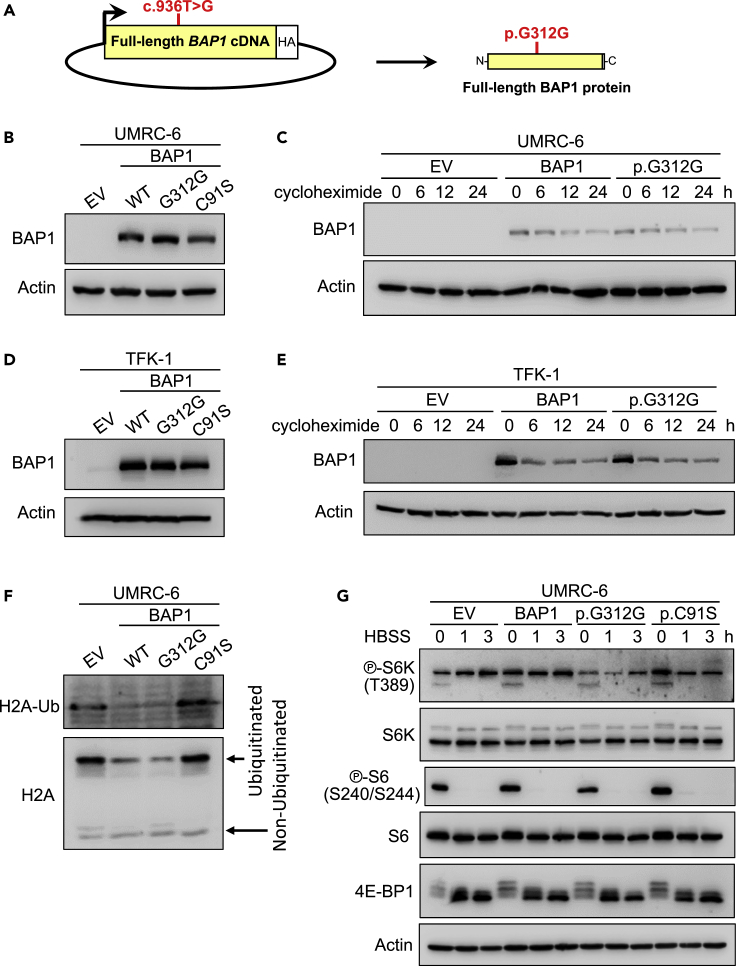
(A) Schema depicting the c.936T>G mutation in the full-length BAP1 cDNA.
(B) UMRC-6 cells stably expressing the p.G312G BAP1 mutant show by western blot similar total protein levels as cells reconstituted with wild-type (WT) BAP1 or the p.C91S mutant. EV, empty vector.
(C) The decay over time of the protein encoded by the cDNA of the synonymous BAP1 mutation is similar to the protein decay from the wild-type BAP1 cDNA in UMRC-6 cells treated with 80 μg/ml of cycloheximide.
(D) BAP1-deficient cholangiocarcinoma TFK-1 cells reconstituted with the p.G312G BAP1 mutant show similar levels of protein expression as wild-type or the p.C91S mutant BAP1.
(E) Western blot of the same stable TFK-1 cell lines treated with 10 μg/ml of cycloheximide for the indicated time shows similar protein decay for the p.G312G mutation as the wild-type BAP1.
(F) Reconstitution of UMRC-6 cells with the p.G312G mutation in BAP1 decreases the ubiquitination of histone H2A to a similar level as to wild-type BAP1, but different than the p.C91S mutant or empty vector.
(G) UMRC-6 cells reconstituted with the p.G312G mutation show similar mTORC1 activation readouts in response to starvation as wild-type and the p.C91S mutant BAP1.