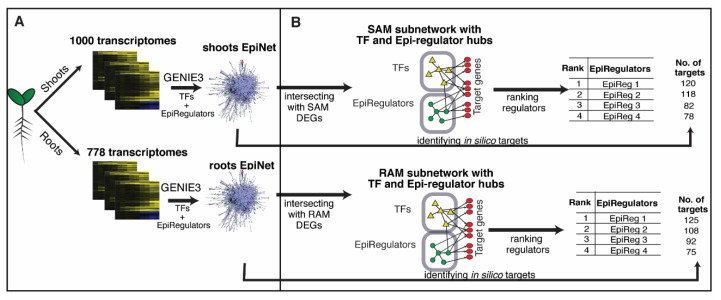
Figure 1.
Workflow showing the analysis steps of this study: (A) 1000 transcriptomes from Arabidopsis shoots and 778 transcriptomes from Arabidopsis roots were downloaded from NCBI’s SRA (short read archive) database. GENIE3 was applied to construct gene regulatory networks from these transcriptomes for shoots and roots, separately, using both transcription factors (TFs) and epigenetic regulators (EpiRegulators) as predictors of gene expression levels. The resultant gene regulatory networks are named EpiNet, for shoots and roots, separately. (B) EpiNet was intersected with differentially expressed genes (DEGs) identified from individual transcriptomic studies to generate subnetworks relevant to the question of interest—for example, shoot apical meristem (SAM) and root apical meristem (RAM) development. The epigenetic regulators involved in controlling the subnetwork can then be ranked based on their influence on the genes in the subnetwork, to identify the most essential epigenetic regulators for the biological process of interest. For each epigenetic regulator, the in silico predicted targets can be identified in the EpiNet.