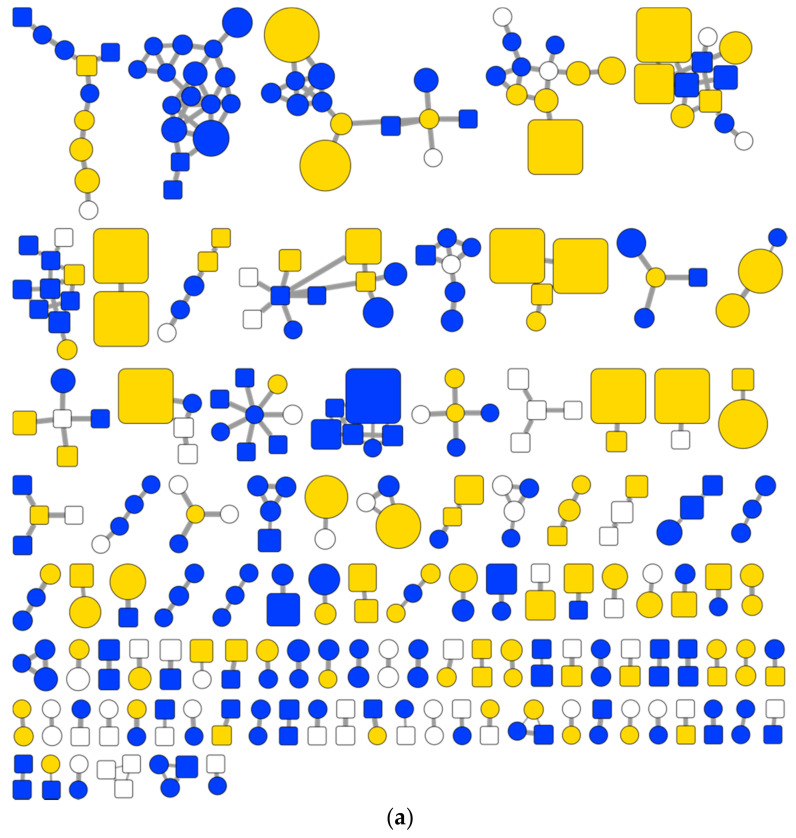
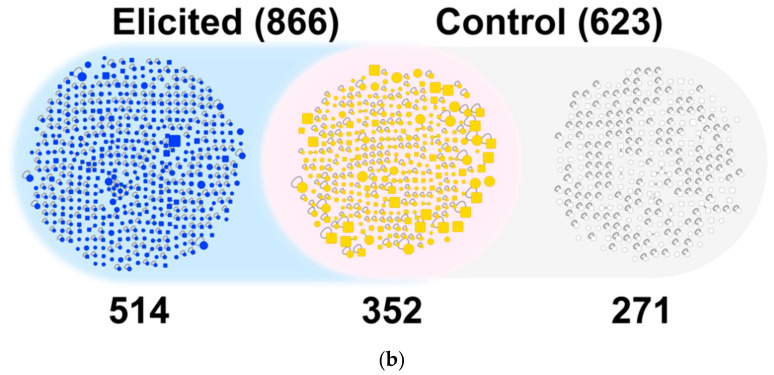
Figure 4.
SSMN analysis of the [M + H]+ consensus spectra linked to bacterial metabolites detected in the EPfB chemical library shows the increment of chemical diversity promoted by bacterial cultivation in the presence of chemical elicitors. The network nodes (circles and squares) represent the [M + H]+ consensus spectra and the links connecting two nodes represent their spectra similarity score, ranging from 0 (totally dissimilar) to 1 (completely identical). The network is organized in spectra clusters by applying a spectra similarity threshold greater or equal to 0.6, limiting the number of neighbors of each node to 15 and limiting the clusters size to 200 nodes. (a) Network organization by the size of the spectra clusters, highlighting the larger clusters at the top of the network. The percent of ion contribution (peak area) of each growth condition to each node is denoted by the size of the node. Nodes are colored according to the sample of origin: exclusive to elicited growth conditions (blue), exclusive to control growth conditions (no elicitor added—in white), and common to both elicited and control conditions (yellow). The link’s width is proportional to the spectra similarity value between the two connected nodes. Self-loops (nodes with no connections) were excluded for better visualization. Nodes were further compared to MS/MS databases and annotated. The identified nodes are shown in squares. (b) Network organization by grouping the nodes detected preferentially under a given growth condition. Numbers indicate the total number of nodes in each group (including self-loops). The same color code in (a) is used.