Figure 5.
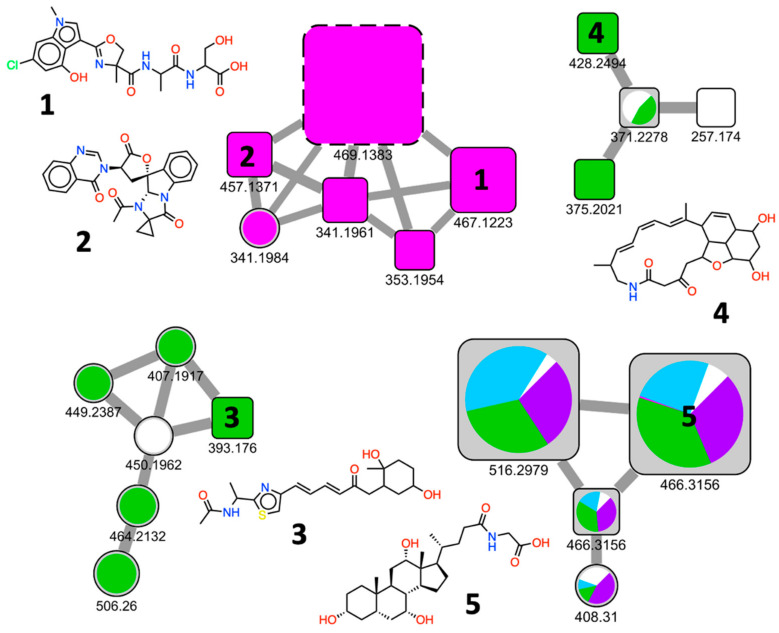
Chemical elicitation induced the production of rare and bioactive bacterial secondary metabolites. Selected clusters of consensus spectra were extracted from the SSMN of the EPfB library. JBIR-35 (1, m/z 467.1223) and tryptoquivaline K (2, m/z 457.1371) were coherently identified from a six-membered cluster elicited by kanamycin. In the same cluster, the major compound m/z 469.1383 (dashed lines) was denoted by a clear chlorine isotope pattern, incompatible with the proposed compound in the database, its identification being ignored from now on. Bacterial compounds leinamycin (LNM) K-3 (3, m/z 393.1760) and verticilactam (4, m/z 428.2494) are also highlighted in two clusters composed mainly by [M + H]+ consensus spectra derived from the EDTA elicited conditions. Glycocholic acid (5, m/z 466.3156), a compound found in marine bacteria, was found comprising a cluster of cholic acid derivatives in most of the growth conditions evaluated. Identified nodes are represented in squares; relative peak areas are represented in the pie-chart, colored according to the elicited growth condition: ampicillin (blue), kanamycin (magenta), procaine (purple), sodium butyrate (red), EDTA (green) and control conditions (white).