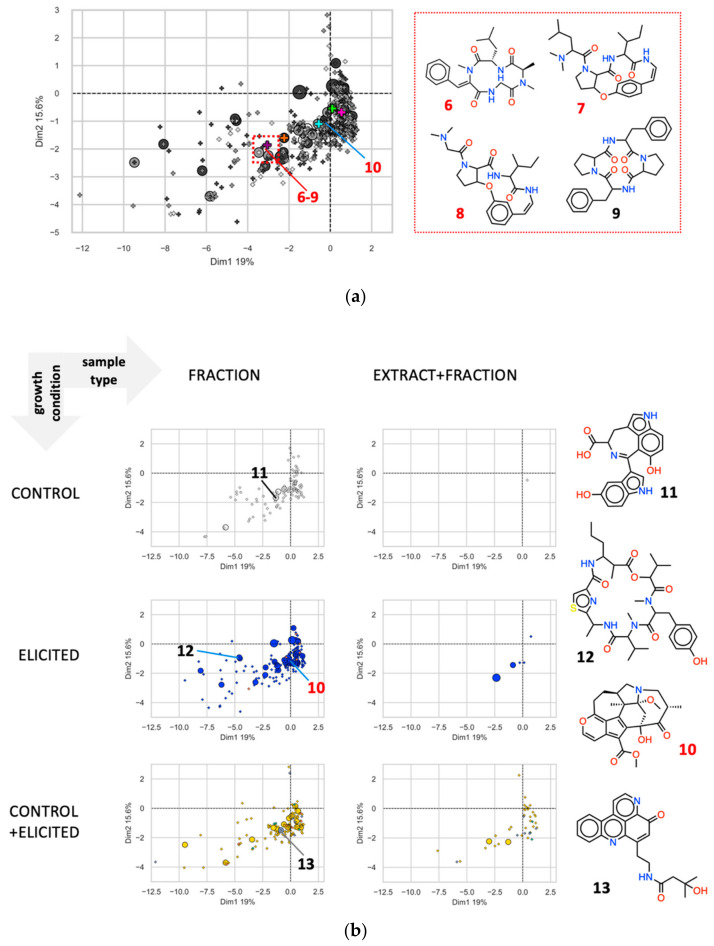
Figure 7.
Expansion of the chemical space by inclusion of similar non-identified spectra and dissection of the chemical space by sample type and growth condition highlights the expression modulation of minor nitrogenated metabolites (Q3) and analogues by chemical elicitors. (a) A principal component analysis was performed based on molecular descriptors of identified [M + H]+ consensus spectra detected in this work. Each identified compound is represented by a cross in the plots and the circles denote the occurrence of non-identified analogous [M + H]+ consensus spectra in the collection. Circle size corresponds to the number of non-identified neighbors connected to the identified compound in the SSMN. Compounds 1–5 are highlighted purple, orange, cyan, green and magenta. (b) The PCA plot was split based on the cultivation method employed for bacterial cultures (growth condition: control (white), elicited (blue) and both (yellow)), and further by the sample type in which the spectra were found (sample type: chromatographic fraction (minor compounds) or both (major compounds)). Compounds derived from bioactive fractions are highlighted in grey (LAMA915), green (LAMA639) and red (B002_754). Representative compounds from the spectra databases used for metabolite identification, sharing spectra similarity to the EPfB [M + H]+ consensus spectra, were selected and are represented; 6: tentoxin; 7: mucronine J; 8: nummularine F; 9: cyclic tetrapeptide (PheProPhePro); 10: daphnicyclidin; 11: hyrtiazepine; 12: ulongamide D; and 13: cystodytin C. Numbers in red indicate natural products reported from organisms other than bacteria.