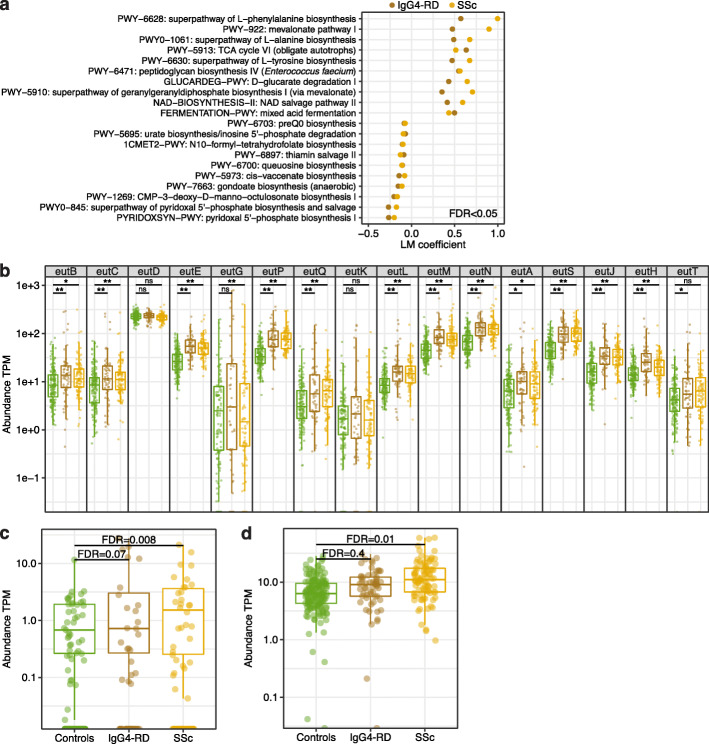
Fig. 4.
Functional microbiome signatures common to IgG4-RD and SSc. a Top 20 significantly up- and downregulated MetaCyc pathways common to IgG4-RD and SSc compared to healthy controls (FDR < 0.05). b Numerous genes belonging to the ethanolamine utilization pathway were increased in abundance in disease. Asterisks (*, **) indicate differential abundance at FDR < 0.2 and FDR < 0.05 levels. Differential abundances (transcripts per million, TPM) of c Sfb1 (SSc FDR = 0.008, IgG4-RD FDR = 0.07) and d HypD (SSc FDR = 0.01) in IgG4-RD and SSc compared to controls. Boxplots show median and lower/upper quartiles; whiskers show inner fences. Full list of differentially abundant MetaCyc pathways and KEGG KO genes are in Additional file 1: Tables S7 and S9