In this study, Sinturel et al. sought to show that the suprachiasmatic nucleus (SCN) synchronizes peripheral circadian oscillators. By using long-term bioluminescence recordings in freely moving mice, they show that the SCN is indeed required for maintaining synchrony between organs and that circadian oscillations persist in the livers of mice devoid of an SCN or oscillators in cells other than hepatocytes.
Keywords: circadian gene expression, in vivo bioluminescence recording, liver, suprachiasmatic nucleus
Abstract
It has been assumed that the suprachiasmatic nucleus (SCN) synchronizes peripheral circadian oscillators. However, this has never been convincingly shown, since biochemical time series experiments are not feasible in behaviorally arrhythmic animals. By using long-term bioluminescence recording in freely moving mice, we show that the SCN is indeed required for maintaining synchrony between organs. Surprisingly, however, circadian oscillations persist in the livers of mice devoid of an SCN or oscillators in cells other than hepatocytes. Hence, similar to SCN neurons, hepatocytes can maintain phase coherence in the absence of Zeitgeber signals produced by other organs or environmental cycles.
The mammalian circadian timing system is composed of a master pacemaker in the brain's suprachiasmatic nucleus (SCN) and cell-autonomous clocks in most tissues (Nagoshi et al. 2004; Welsh et al. 2004). These oscillators consist of two interlocked transcriptional-translational feedback loops (TTFLs) (Schibler et al. 2015; Cox and Takahashi 2019). In the primary TTFL, Period and Cryptochrome genes (Per1, Per2, Cry1, and Cry2) are activated by CLOCK-BMAL1 heterodimers. PER and CRY proteins then assemble into large repressor complexes that inhibit the CLOCK-BMAL1 activity (Schmalen et al. 2014; Aryal et al. 2017). Consequently, the levels of PER and CRY transcripts and proteins decrease, and a new round of PER and CRY expression can ensue. In a secondary TTFL REV-ERBα,β repressors counteract the stimulation of Bmal1 and Clock transcription by RORα,β,γ activators.
Since circadian clocks can measure a 24-h day only approximately, they must be synchronized daily. The SCN is synchronized primarily by light–dark cycles (Patton and Hastings 2018; Hastings et al. 2019). The strong phase coupling among neurons and between neurons and astrocytes establish circadian phase coherence even in cultured SCN explants (Yamazaki et al. 2000; Liu et al. 2007; Herzog et al. 2017). Peripheral clocks are synchronized by feeding–fasting rhythms (Damiola et al. 2000; Stokkan et al. 2001; Crosby et al. 2019), blood-borne signaling molecules, and body temperature cycles (for review, see Schibler et al. 2015), but they rapidly loose phase coherence in vitro (Welsh et al. 2004). Using a bioluminescence imaging method, Tahara et al. (2012) found that about half of SCN-lesioned animals still displayed rhythms in liver, kidney, and submaxillary gland. However, their method involved anesthesia and is thus not appropriate for continuous recording over multiple days. Here, we adapted the RT-Biolumicorder technique (Saini et al. 2013) to the monitoring of circadian gene expression in freely moving mice for many weeks to examine the impact of the SCN and feeding–fasting cycles on peripheral clocks. Moreover, we monitored circadian gene expression in behaviorally arrhythmic mice harboring clocks exclusively in hepatocytes.
Results and Discussion
To extend the recording time of the original RT-Biolumicorder technology (Saini et al. 2013), we provided luciferin in the drinking water instead of through an Alzet micro-osmotic pump (Fig. 1A–C; Supplemental Fig. S1A–C). The two delivery methods were then compared for the monitoring of whole-body Per2::luciferase expression in hairless mice entrained by a skeleton photoperiod. As shown in Figure 1, A–C, the bioluminescence displayed high-amplitude circadian oscillations with both methods (Fig. 1A). Spontaneous locomotor and drinking activities showed similar temporal profiles in the presence or absence of the pump (Fig. 1B,C; Supplemental Fig. S1D). Likewise, we observed robust circadian bioluminescence cycles with both delivery methods when recording a mouse whose liver was transduced by a Rev-erbα-luciferase adenoviral vector (Supplemental Fig. S1A–C). Importantly, animals maintained in constant darkness and receiving luciferin in the drinking water displayed a nearly antiphasic expression of Rev-erbα-luciferase and Per2::luciferase (Fig. 1D,E) with indistinguishable period length or phase (Fig. 1F,G). Therefore, the drinking rhythms, generated by rest–activity cycles, did not interfere dramatically with the bioluminescence recordings. Fast Fourier Transform (FFT) analysis showed that the amplitude of the FFT peak at 24 h was similar when luciferin was delivered with the drinking water (Supplemental Fig. S1E), and the luciferin delivery method did not significantly affect the accuracy of period length (Supplemental Fig. S1F).
Figure 1.
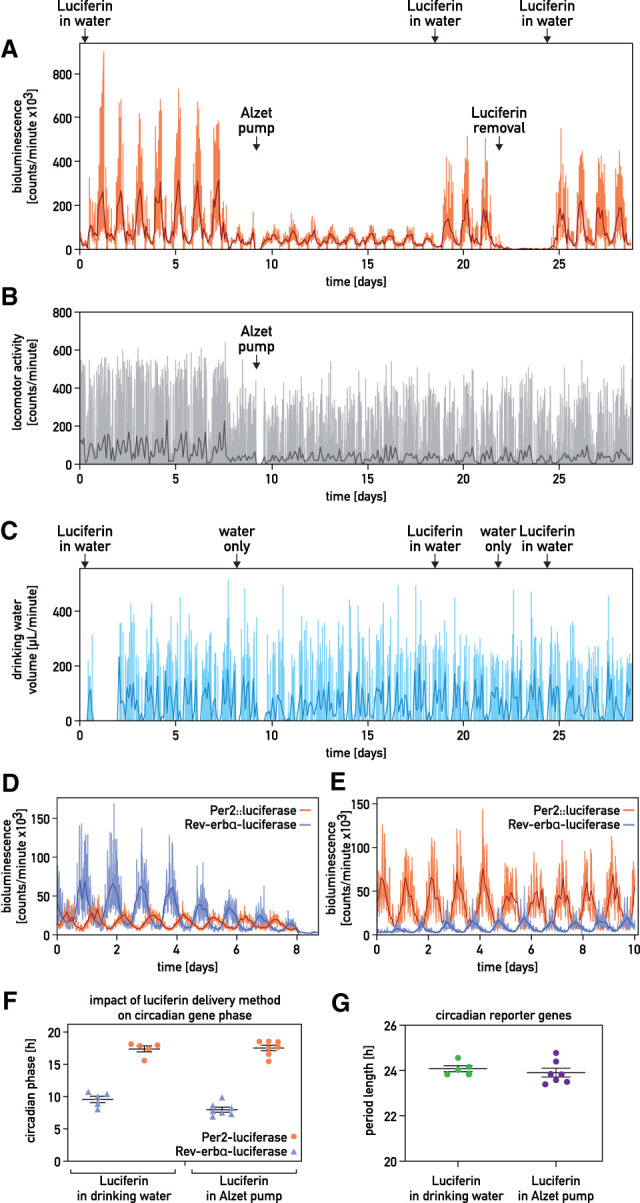
Long-term recording of circadian whole-body bioluminescence. (A–C) Whole-body recording of a hairless Per2::luc mouse entrained by a skeleton photoperiod. Luciferin was delivered as indicated. (A) Bioluminescence. (B) Spontaneous locomotor activity. (C) Drinking activity. (Orange, gray, and light blue lines in A–C) Raw data (1-min photon counts, 1-min activity counts, and 1-min water consumption, respectively), (darker lines in A–C) 120-min time-binned traces. (D,E) Whole-body Per2::luciferase and hepatic Rev-erbα-luciferase recordings from two different animals kept in constant darkness and receiving luciferin through Alzet pumps (D) or with the drinking water (E). (Light lines) Raw data, (darker lines) 120-min time-binned traces. (F,G) Relationship between the phases (F) and the period lengths (G) of Per2::luciferase and hepatic Rev-erbα-luciferase expression of mice receiving luciferin in the drinking water (n = 5) or by an Alzet micro-osmotic pumps (n = 7). Mice were kept in constant darkness.
We next performed experiments with PER2::luciferase reporter mice (Yoo et al. 2004) whose SCN was intact or ablated by stereotaxic lesions (Fig. 2A,B; Supplemental Fig. S2A–F). As expected, locomotor activity was lost in SCN-lesioned but not in sham-operated animals kept in constant darkness (D/D) (Fig. 2C; Supplemental Fig. S3). To examine the impact of feeding–fasting rhythms on locomotor activity and bioluminescence, PER2::luc mice were maintained in D/D and fed according to the regimens indicated on top of Figure 2A. While the locomotor activity of SCN-lesioned mice was arrhythmic in animals fed ad libitum, it showed clear rhythmicity in animals subjected to 12 h/12 h feeding–fasting rhythms (Fig. 2D–G). As shown in Figure 2A and Supplemental Figure S2C,D, whole-body bioluminescence (i.e., the sum of photons emitted by many organs) showed high-amplitude oscillations in sham-operated mice. In contrast, these rhythms were strongly dampened in SCN-lesioned mice, suggesting that the SCN is required for the synchronization of oscillators among different tissues. Imposing feeding rhythms synchronized PER2::luc expression in both sham-operated and SCN-lesioned mice (Fig. 2A,B,E; Supplemental Fig. S2C–F). In agreement with previous studies (Damiola et al. 2000; Le Minh et al. 2001; Saini et al. 2013), the phase of circadian gene expression was shifted more rapidly in SCN-lesioned as compared with sham-operated mice (Supplemental Fig. S2G). The period lengths and amplitudes of the FFT peaks at 24 h were similar in SCN-proficient and -deficient mice when animals were subjected to feeding–fasting cycles (Fig. 2F–I). Unexpectedly, the overall bioluminescence was markedly higher in recordings of SCN-lesioned as compared with sham-operated mice (Supplemental Fig. S2H). Therefore, signals depending directly or indirectly on the SCN appeared to reduce, rather than increase, the magnitude of PER2::luciferase expression in peripheral tissues.
Figure 2.
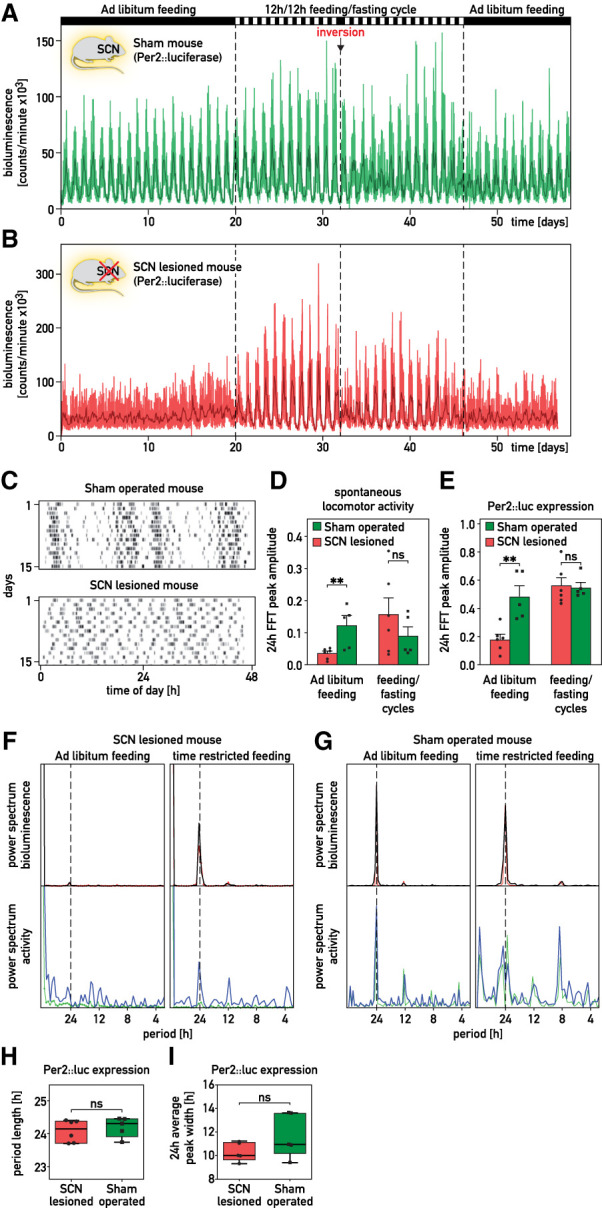
The SCN is required for the synchronization of peripheral clocks among organs. (A,B) Whole-body bioluminescence for sham-operated (A) and SCN-lesioned (B) Per2::luc mice kept in constant darkness. The feeding regimens are reported above the graphs. The spontaneous locomotor activities of the animals are shown in Supplemental Figure S2A,B. (Thin lines) Raw data (1 min photon counts), (bold lines) 120-min time-binned traces. (C) Actograms of spontaneous locomotor activity of the sham-operated mouse used in A and the SCN-lesioned mouse used in B. After surgery, mice were housed in 12-h/12-h light/dark (LD) cycles and then recorded during 15 d in constant darkness (D/D). (D,E) Amplitude of the FFT peak in the circadian range (≈24 h) of locomotor activities (D) and of Per2::luciferase expression (E) of SCN-lesioned and sham-operated mice subjected to different feeding regimens (means ± SEM, n ≥ 5 per group). (F,G) Periodograms (FFT analysis) of bioluminescence and locomotor activity recorded for the sham-operated mouse displayed in A and the SCN-lesioned mouse displayed in B. (Red and green lines) Raw data (1-min counts), (black and blue lines) integrated data over 120 points. (H,I) Period length from FFT analysis (H) and average oscillation width (I) in sham-operated and SCN-lesioned animals subjected to feeding–fasting cycles. Whiskers of the box plots range from the minimum to the maximum values. The median values are indicated by the horizontal segment within the box, n ≥ 5 per group. Statistical tests for D, E, H, and I were two-tailed Mann–Whitney test. (**) P < 0.01, (ns) nonsignificant.
Since a residual rhythmicity of PER2::luc expression could still be discerned in SCN-lesioned mice, we suspected that some cellular oscillators could still be in phase in these animals. To examine this conjecture for liver, we transduced liver cells of sham-operated and SCN-lesioned mice with the adenoviral Rev-erbα-luciferase vectors via tail vein injection (Fig. 3A,B; Supplemental Fig. S4A–E; Saini et al. 2013). Hepatic Rev-erbα-luciferase expression oscillated robustly in both SCN-proficient and -deficient mice monitored in constant condition, irrespective of whether luciferin was administered through the drinking water (Fig. 3B) or via an Alzet pump (Supplemental Fig. S4B–E). A similar observation was made for hepatic Bmal1-luciferase expression (Supplemental Fig. S4E–G). The FFT analysis of the bioluminescence profiles (Fig. 3C,D) revealed no statistically significant differences for the FFT peak at 24 h in SCN-proficient and -deficient animals (Fig. 3E). As expected, the SCN-lesioned mice displayed significantly lower amplitudes of the FFT peak at 24 h for the locomotor activity (Fig. 3F). Although hepatic Rev-erbα-luciferase expression in SCN-lesioned mice showed a somewhat larger variability of the period length and a slightly increased width of the 24-h oscillations (Fig. 3G,H), these differences were not statistically significant. As the amplitude and peak width were not significantly different between sham-operated and SCN-lesioned mice fed ad libitum or subjected to a feeding–fasting regimen (Fig. 3E,H), the SCN did not markedly enhance the amplitude of hepatocyte oscillators and/or their intercellular coupling within the liver. Given that the efficiency of adenoviral transductions varied somewhat between animals, we could not directly compare the magnitude of bioluminescence generated by the livers of sham-operated and SCN-lesioned mice.
Figure 3.
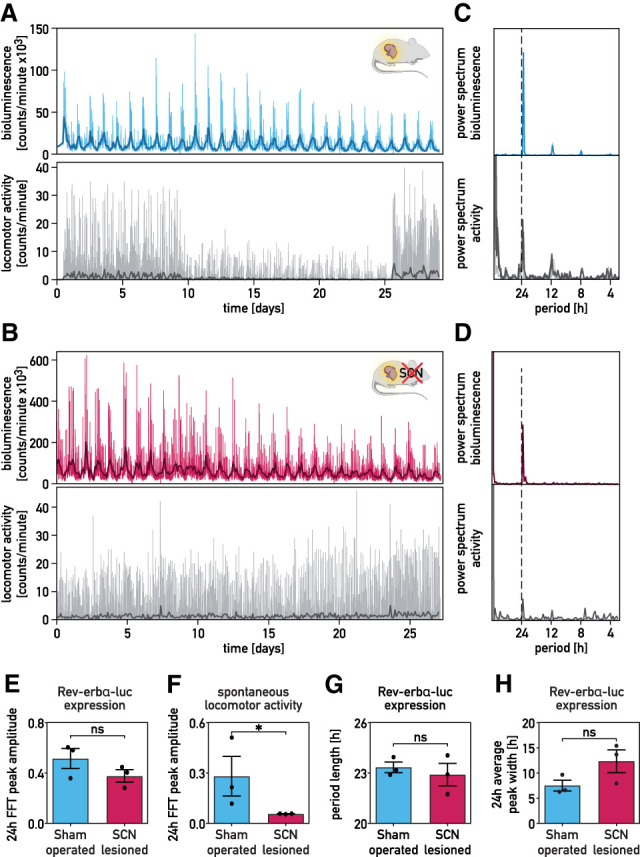
Hepatocyte oscillators maintain synchrony in SCN-lesioned animals. (A,B) Recordings of circadian Rev-erbα-luciferase expression in the livers of sham-operated (blue; A) and SCN-lesioned (pink; B) mice receiving luciferin in the drinking water. Mice transduced with the Adv-Rev-erbα-luc vector were shaved in a dorsal region over the liver. Forty-eight hours after transduction, bioluminescence was monitored in constant darkness (food and water ad libitum). Spontaneous locomotor activity profiles monitored simultaneously are shown below the bioluminescence tracings. The drop of the signal for locomotor activity in the middle of the recording was caused by the obstruction of the IR detector by litter piled up by the mouse. (Thin colored and gray lines ) Raw data (1-min counts), (bold lines) 120-min time-binned traces. (C,D) Periodograms (FFT analysis) of the data shown in A and B. (Thin colored and gray lines) Raw data (1-min counts), (bold lines) 120-min time-binned traces. (E,F) Amplitudes of the FFT peaks in the circadian range of hepatic Rev-erbα-luciferase expression (E) and spontaneous locomotor activity rhythms (F) of SCN-lesioned and sham-operated mice maintained in constant conditions (means ± SEM, n = 3 per group). The statistical test was a one-tailed Mann–Whitney test. (*) P < 0.05, (ns) nonsignificant. (G,H) Period lengths of Rev-erbα-luciferase expression (G) and average oscillation width in the circadian range (≈24 h) (H) of sham-operated and SCN-lesioned animals maintained in constant conditions (means ± SEM, n = 3 per group). The statistical test was a two-tailed Mann–Whitney test. (ns) Nonsignificant.
To examine whether liver cells remain synchronized in the absence of nonhepatic signals, we studied mice (kept in constant darkness) harboring clocks exclusively in hepatocytes. These animals, dubbed “hepatocyte clock-only mice,” contain a Bmal1-allele carrying a lox-stop signal between exon 5 and 6 that can be deleted with a Cre recombinase specifically expressed in hepatocytes (Fig. 4A; Supplemental Fig. S5A,B). In parallel, we used Bmal1+/− mice, in which Bmal1 is expressed throughout the body, and arrhythmic Bmal1−/− mice. As shown in Figure 4B,C and Supplemental Figure S5C–K, bioluminescence recordings of Bmal1+/− and Bmal1−/− mice revealed circadian and arrhythmic Rev-erbα-luciferase expression, respectively, in the livers of these animals. Strikingly, we observed rhythmic oscillations of hepatic Rev-erbα-luciferase expression in “hepatocyte clock-only mice,” but, as expected, not in Bmal1−/− mice devoid of a Cre recombinase (Fig. 4D). “Hepatocyte clock-only mice” showed less stable rhythms of Rev-erbα-luciferase expression than Bmal1+/− or SCN-lesioned animals. However, these oscillations were statistically highly significant (Fig. 4E–G) and revealed a dominant peak at around 24 h in all periodograms (Supplemental Fig. S5L–P). As anticipated, neither Bmal1−/−mice nor “hepatocyte clock-only mice” showed behavioral rhythms (Fig. 4H). Hence, hepatocytes were partially synchronized in behaviorally arrhythmic mice that harbor circadian oscillators exclusively in hepatocytes.
Figure 4.
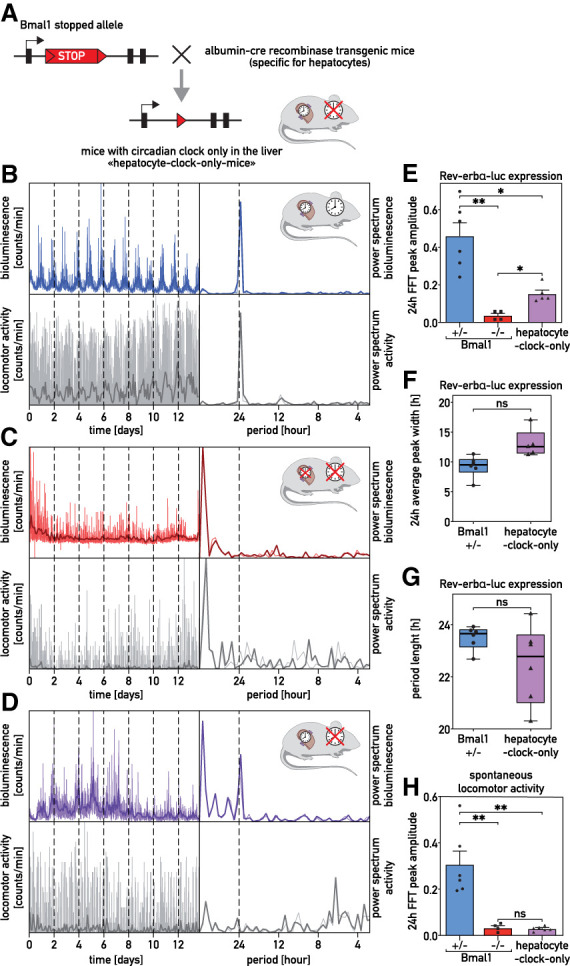
The livers of “hepatocyte clock-only mice” keep partial phase coherence. (A) Scheme of the lox-stop allele used to generate “hepatocyte clock-only mice.” (B–D) Recordings of hepatic expression of Rev-erbα-luciferase in Bmal1+/− mice (blue; B), Bmal1−/− mice (red; C), and “hepatocyte clock-only mice” (violet; D) receiving luciferin in the drinking water. Mice were transduced with the Adv-Rev-erbα-luc expression vector, and bioluminescence was recorded under constant conditions (D/D, food and water ad libitum). Spontaneous locomotor activity profiles are shown below the bioluminescence tracings. The FFT analysis of the bioluminescence and locomotor activity recordings are represented at the right of the bioluminescence and activity tracings. (Thin colored and gray lines) Raw data (1-min counts), (bold lines) 120-min time-binned traces. (E) Amplitude of the FFT peaks in the circadian range, determined for hepatic Rev-erbα-luciferase expression in Bmal1+/− mice, Bmal1−/−mice, and “hepatocyte clock-only mice.” (F) Width of the oscillations for hepatic Rev-erbα-luciferase expression in the circadian range (≈24 h) in Bmal1+/− and “hepatocyte clock-only mice.” (G) Period length of Bmal1+/− mice and “hepatocyte clock-only mice.” (H) Amplitude of the FFT peaks in the circadian range, determined for the spontaneous locomotor activity rhythms of Bmal1+/− mice, Bmal1−/− mice, and “hepatocyte clock-only mice.” The statistical test for E and H was a Kruskal–Wallis test corrected by FDR (means ± SEM, n ≥ 4 per group), and for F and G a two-tailed Mann–Whitney test (n ≥ 4 per group). (**) P < 0.01, (*) P < 0.05, (ns) nonsignificant. (F,G) Whiskers of the box plots represent minimum to maximum values with median value indicated by the horizontal segment within the box.
In contrast to previous reports (Guo et al. 2006; Koronowski et al. 2019), our findings show that the SCN is not required for maintaining phase coherence between hepatocytes. Based on photon imaging of anesthetized, SCN-lesioned PER2::luc mice (Tahara et al. 2012) and the bioluminescence recording of tissue explants from SCN-lesioned PER2::luc mice (Yoo et al. 2004), similar conclusions were reached.
In “hepatocyte clock-only mice,” the synchronization of liver clocks was less complete than in SCN-lesioned mice. Conceivably, in SCN-lesioned mice, signals emitted by liver cells other than hepatocytes (or nonhepatic tissues) might have participated in the synchronization of hepatocyte clocks. Indeed, a recent study provided direct evidence for intercellular communication by different cell types in controlling rhythmic liver gene expression (Guan et al. 2020). In contrast to the results reported here, Koronowski et al. (2019) did not observe rhythmic gene expression in “hepatocyte clock-only mice” kept in constant darkness when comparing liver mRNA levels between individual animals. Presumably, the phases of hepatic circadian gene expression were different between these individuals. The data presented here thus exemplify the importance of real-time recording when studying cyclic gene expression in behaviorally arrhythmic animals.
In conclusion, and as illustrated in Figure 5, our study revealed that the SCN master pacemaker is required for keeping phase coherence between different organs in mice kept under constant conditions. However, within the liver, hepatocytes remain perfectly synchronized in the absence of an SCN and maintain some phase coherence even in mice in which circadian oscillators function exclusively in hepatocytes. The identification of signaling pathways involved in the phase coupling between hepatocyte oscillators will be an enticing future endeavor.
Figure 5.
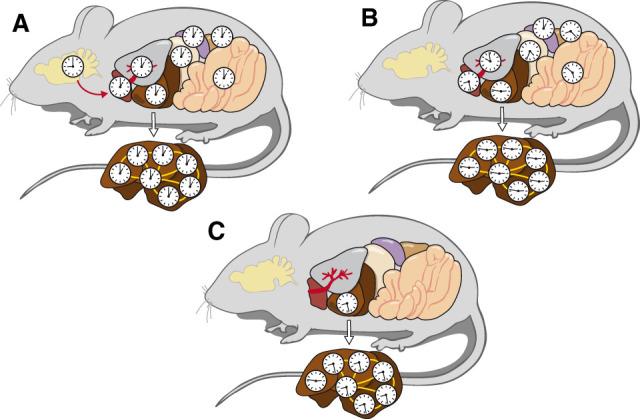
Scheme of circadian hepatocyte clock synchrony in control animals and in absence of the SCN or other extrahepatic clocks. (A) In intact, behaviorally rhythmic mice, the central pacemaker in the SCN synchronizes circadian oscillators to similar phases in peripheral organs (red arrow). The phases of peripheral clocks are somewhat delayed with regard to the phase of the SCN (Damiola et al. 2000). (B) In SCN-lesioned, behaviorally arrhythmic mice, the circadian clocks of different peripheral organs have different phases, but within the liver the cellular oscillators remain synchronized. (C) In behaviorally arrhythmic “hepatocyte clock-only mice,” a large proportion of hepatocyte clocks are still in phase. The findings illustrated in B and C suggest that circadian hepatocyte oscillators are phase coupled, as indicated by the yellow lines connecting hepatocyte clocks.
Materials and methods
Animal care
Male hairless mice (strain code 477; formerly SKH1 mice with strain code 313) were purchased from Charles River, and PER2::Luc mice were generously provided by Joseph S. Takahashi (University of Texas Medical Center, Dallas, TX) (Yoo et al. 2004). Hairless PER2::Luc knock-in mice were generated by crossing SKH1 mice with PER2::Luc mice. C57BL/6J mice homozygous for the conditional Bmal1 stopped allele (Bmal1 “stopped” mice) (Petrenko et al. 2020) were bred with mice carrying a Cre recombinase transgene under the control of the albumin promoter [albumin-Cre, B6.Cg-Tg(Alb-cre)21Mgn/J] (Postic et al. 1999), generating “hepatocyte clock-only mice” with Bmal1 expression restricted to the hepatocytes (for details, see the Supplemental Material). Bmal1−/− (“stopped”) and Bmal1+/− heterozygote littermates were used as controls in the experiments.
All animals were housed according to Swiss standard conditions. All the experiments were performed on healthy mice, with normal immune status, and approved by the Veterinary Offices of the Cantons of Geneva and Vaud. When indicated, feeding or light–dark cycles were imposed during bioluminescence recording. Entrainment with skeleton photoperiods (Fig. 1; Supplemental Fig. S1) was performed as described previously with two light pulses of 30 min at 12 h intervals (Friedrich and Soriano 1991; Oishi et al. 2002).
SCN ablation
Bilateral lesion of the SCN was performed stereotaxically (Kopf Instruments 963LS) under ketamine/xylazine anesthesia (intraperitoneal injection, 75 and 10 mg/kg, at a volume of 8 mL/kg) as described (Saini et al. 2013). The success of the experiments was controlled by recording spontaneous locomotor activity in individual cages equipped with infrared sensors (Visonic) in free-running conditions (constant darkness, food and water provided ad libitum) for at least 10 d.
Generation of circadian reporter luciferase-expressing adenoviral expression vectors
Rev-erbα-luciferase and Bmal1-luciferase-expressing adenoviral constructs have been previously described (Kreppel et al. 2002; Saini et al. 2013).
Luciferin delivery and tail vein injection
For the experiments with micro-osmotic pumps, pumps (model 2001, blue flow moderator 0002489, Alzet, Charles River) were filled with 90 mg/mL luciferin (306A NF-FF free-acid firefly D-Luciferin, Nanolight Technology, Prolume Ltd.) and implanted into the intraperitoneal cavity under anesthesia, as described previously (Saini et al. 2013). For long-term recordings, luciferin (XenoLight D-Luciferin, potassium salt; PerkinElmer, 4.9 mM stock concentration in sterile double-distilled water) was diluted in tap water at a final concentration of 1.56 mg/mL and provided in the drinking water. Delivery of adenoviral-luciferase reporters (2.5 × 1010 to 1 × 1011 adenoviral particles) into the liver was performed as described previously (Saini et al. 2013). After 40–48 h of recovery, the animals were transferred to the RT-Biolumicorder, and the bioluminescence monitoring started.
RT-Biolumicorder measurements and data analysis
The RT-Biolumicorder (Lesa-Technology SA) has been designed and engineered as described in Saini et al. (2013), and details are in the Supplemental Material.
Original data of bioluminescence, activity, and drinking temporal profiles were collected at 1-min intervals (raw data are in Supplemental Table S1) and analyzed using the software Igor Pro v. 8.0.4 (WaveMetrics, Inc.). A custom function was applied to bin the data points summing their values over 120-min intervals. This binning time procedure was especially required for analyzing the activity traces, which might contain multiple voids. Igor Pro built-in function DSP Periodogram was applied on each binned trace to compute the power spectrum
where F indicates the Fast Fourier Transform operator, and N is the total number of samples. The data sets were automatically detrended prior to undergoing FFT. The power spectra were successively normalized by the sum of their values. An automatic procedure was applied to fit with a Gaussian curve the power spectrum in an interval centered around the 24-h period. The fitted values for the Gaussian peak amplitude and central position were used for the statistical comparisons among different data sets. Note that the fitted values were retained for the analysis only if the circadian peak at 24 h resulted statistically significant. To determine the width of 24-h oscillations, we applied a Gaussian multipeak fit procedure to the complete time traces and successively calculated the average peak FWHM and standard deviation for each condition. For the calculation of phase angle changes as a function of time after inversion of the feeding regimen, the peaks in the oscillatory bioluminescence response of a given animal were fitted with a Gaussian function to automatically retrieve the positions of their maxima. The time shift with respect to an ideal 24-h daily cycle was computed by subtracting to the nth peak position, the position of the first peak, and the total number of daily cycles. (n − 1) × 24 h. The phase shift was finally calculated by setting 24 h = 360°.
Statistical analyses
For comparison across different experimental groups, statistical significance was assessed with statistical tests in Prism 8 (GraphPad). The statistical test used, the significance values, and the number of experimental replicates for each experimental group are reported in the figure legends. Grouped data were plotted in Prism 8 as mean ± SEM.
RNA analysis by quantitative real-time RT-PCR
Whole-cell RNAs from liver, kidney, spleen, and brown adipose tissue (BAT) were purified as previously described (Sinturel et al. 2017; details in the Supplemental Material).
For the quantification of specific transcripts, cDNAs were synthesized from total RNAs using random hexamers and SuperScript II reverse transcriptase (Thermo Fisher Scientific) following the supplier's instructions. The cDNAs were quantified by real-time PCR-amplification in a LightCycler 480 (Roche), using the SYBR Green Master Mix (Roche). Mean levels were calculated from triplicate PCR assays for each sample and normalized to the amounts of Gapdh transcripts (Supplemental Table S2).
Competing interest statement
A.L. is a stockholder of Lesa-Technology SA, which makes the RT-Biolumicorder commercially available.
Supplementary Material
Acknowledgments
We thank M. Stoffel, ETH Zürich, for providing Albumin Cre mice; J. Takahashi, University of Texas Southwestern Medical Center, Dallas, for providing PER2::luc mice; and N. Roggli for the artwork. Work in the laboratory of U.S. was supported by the Swiss National Science Foundation (SNSF; grants 31-113565 and 31-128656/1), the European Research Council (ERC-2009-AdG-TIMESIGNAL-250117), the State of Geneva, and the Louis Jeantet Foundation of Medicine. F.S. was supported by a Swiss Society of Endocrinology and Diabetology (SGED) Young Investigator grant. Work in the laboratory of C.D. was supported by SNSF grants 31003A_166700/1 and 310030_184708/1, the Vontobel Foundation, the Novartis Consumer Health Foundation, the Swiss Life Foundation, the European Foundation for the Study of Diabetes/Novo Nordisk Program for Diabetes Research in Europe, and the Olga Mayenfisch Foundation. Work in the laboratory of C.W. was supported by a grant from the National Institutes of Health (NS055831) and a fellowship to D.K. from the SNSF. Work in the laboratory of P.F. was supported by SNSF grant 31003A_146694 and the State of Vaud. Work in the laboratory of F.K. and C.H. was supported by the Center of Biomedical Research and Education (ZBAF) at Witten/Herdecke University. L.B.’s work was supported by the State of Geneva.
Author contributions: F.S., C.D., P.F., and U.S. designed the experiments. F.S., P.G., and V.P. did the mouse husbandry and performed the RT-Biolumicorder recording and biochemical experiments. F.K. and C.H. produced the adenoviral vectors. A.L. engineered and constructed the RT-Biolumicorder. K.F.S., D.K., and C.W. produced the Bmal1-lox-stop mice. Y.E. and P.F. performed the SCN lesions. L.B. processed the RT-Biolumicorder data. F.S., C.D., and U.S. wrote the first draft of the manuscript that was edited and approved by all coauthors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.346460.120.
Freely available online through the Genes & Development Open Access option.
References
- Aryal RP, Kwak PB, Tamayo AG, Gebert M, Chiu PL, Walz T, Weitz CJ. 2017. Macromolecular assemblies of the mammalian circadian clock. Mol Cell 67: 770–782.e6. 10.1016/j.molcel.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Takahashi JS. 2019. Circadian clock genes and the transcriptional architecture of the clock mechanism. J Mol Endocrinol 63: R93–R102. 10.1530/JME-19-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby P, Hamnett R, Putker M, Hoyle NP, Reed M, Karam CJ, Maywood ES, Stangherlin A, Chesham JE, Hayter EA, et al. 2019. Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell 177: 896–909.e20. 10.1016/j.cell.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961. 10.1101/gad.183500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev 5: 1513–1523. 10.1101/gad.5.9.1513 [DOI] [PubMed] [Google Scholar]
- Guan D, Xiong Y, Trinh TM, Xiao Y, Hu W, Jiang C, Dierickx P, Jang C, Rabinowitz JD, Lazar MA. 2020. The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science 369: 1388–1394. 10.1126/science.aba8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Lehman MN, Bittman EL. 2006. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: effects of transplanting the pacemaker. J Neurosci 26: 6406–6412. 10.1523/JNEUROSCI.4676-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, Brancaccio M. 2019. The mammalian circadian timing system and the suprachiasmatic nucleus as its pacemaker. Biology (Basel) 8: 13. 10.3390/biology8010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH. 2017. Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb Perspect Biol 9: a027706. 10.1101/cshperspect.a027706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronowski KB, Kinouchi K, Welz PS, Smith JG, Zinna VM, Shi J, Samad M, Chen S, Magnan CN, Kinchen JM, et al. 2019. Defining the independence of the liver circadian clock. Cell 177: 1448–1462.e14. 10.1016/j.cell.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreppel F, Biermann V, Kochanek S, Schiedner G. 2002. A DNA-based method to assay total and infectious particle contents and helper virus contamination in high-capacity adenoviral vector preparations. Hum Gene Ther 13: 1151–1156. 10.1089/104303402320138934 [DOI] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schütz G, Schibler U. 2001. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. Embo j 20: 7128–7136. 10.1093/emboj/20.24.7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129: 605–616. 10.1016/j.cell.2007.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. 2004. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705. 10.1016/j.cell.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Oishi K, Fukui H, Sakamoto K, Miyazaki K, Kobayashi H, Ishida N. 2002. Differential expressions of mPer1 and mPer2 mRNAs under a skeleton photoperiod and a complete light-dark cycle. Brain Res Mol Brain Res 109: 11–17. 10.1016/S0169-328X(02)00457-6 [DOI] [PubMed] [Google Scholar]
- Patton AP, Hastings MH. 2018. The suprachiasmatic nucleus. Curr Biol 28: R816–R822. 10.1016/j.cub.2018.06.052 [DOI] [PubMed] [Google Scholar]
- Petrenko V, Stolovich-Rain M, Vandereycken B, Giovannoni L, Storch KF, Dor Y, Chera S, Dibner C. 2020. The core clock transcription factor BMAL1 drives circadian β-cell proliferation during compensatory regeneration of the endocrine pancreas. Genes Dev 34: 1650–1665. 10.1101/gad.343137.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274: 305–315. 10.1074/jbc.274.1.305 [DOI] [PubMed] [Google Scholar]
- Saini C, Liani A, Curie T, Gos P, Kreppel F, Emmenegger Y, Bonacina L, Wolf JP, Poget YA, Franken P, et al. 2013. Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev 27: 1526–1536. 10.1101/gad.221374.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Gotic I, Saini C, Gos P, Curie T, Emmenegger Y, Sinturel F, Gosselin P, Gerber A, Fleury-Olela F, et al. 2015. Clock-Talk: interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb Symp Quant Biol 80: 223–232. 10.1101/sqb.2015.80.027490 [DOI] [PubMed] [Google Scholar]
- Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, Prabu JR, Benda C, Kramer A, Wolf E. 2014. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157: 1203–1215. 10.1016/j.cell.2014.03.057 [DOI] [PubMed] [Google Scholar]
- Sinturel F, Gerber A, Mauvoisin D, Wang J, Gatfield D, Stubblefield JJ, Green CB, Gachon F, Schibler U. 2017. Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169: 651–663.e14. 10.1016/j.cell.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. 2001. Entrainment of the circadian clock in the liver by feeding. Science 291: 490–493. 10.1126/science.291.5503.490 [DOI] [PubMed] [Google Scholar]
- Tahara Y, Kuroda H, Saito K, Nakajima Y, Kubo Y, Ohnishi N, Seo Y, Otsuka M, Fuse Y, Ohura Y, et al. 2012. In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol 22: 1029–1034. 10.1016/j.cub.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. 2004. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14: 2289–2295. 10.1016/j.cub.2004.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. 2000. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288: 682–685. 10.1126/science.288.5466.682 [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101: 5339–5346. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


