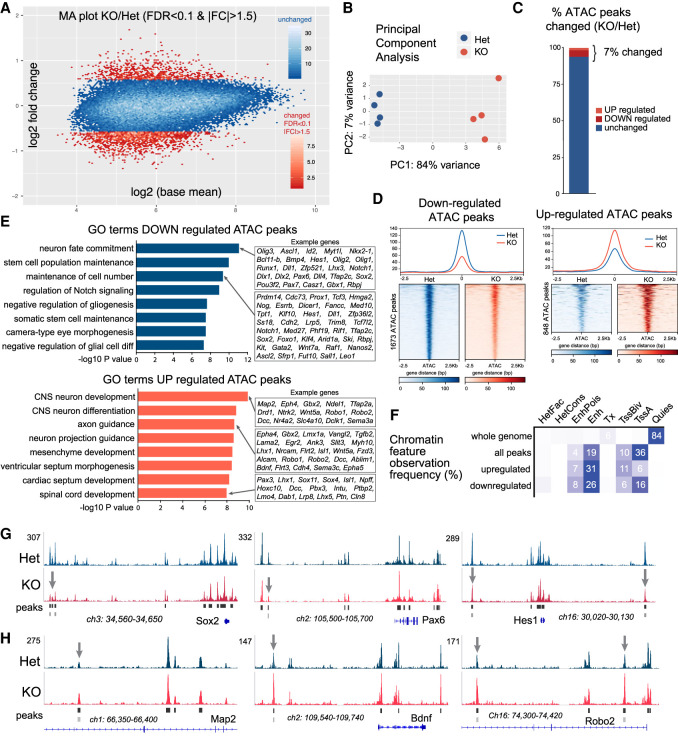
Figure 6.
BAF53a regulates chromatin accessibility at NSPC identity and neural differentiation genes. (A) Genome-wide chromatin accessibility changes as measured by ATAC-seq in BAF53a mutant (Nestin-Cre;Baf53afl/−, KO) and heterozygous control (Nestin-Cre;Baf53afl/+, Het) forebrains at E15.5 (n = 4). Magnitude and average (MA) plot (KO/Het) of ATAC-seq data, displaying significantly changed peaks in red (FDR < 0.1 and fold change > 1.5). (B) PCA plot highlighting reproducibility of biological replicates that cluster according to genotypes. (C) Percentage of the total 39,041 ATAC peaks that are up-regulated or down-regulated. (D) Genome-wide accessibility profiles at up-regulated and down-regulated ATAC peaks in BAF53a KO and Het forebrains. (E) Analysis of gene ontology (GO) terms of biological processes associated with the genes that display gains or losses in accessibility in regulatory elements that control their expression. GREAT software was used to associate ATAC peak with a given gene. (F) Chromatin feature analysis reveals frequency of accessibility changes (percentage) according to specific regulatory elements. The peaks that display changes in accessibility are most frequently associated with enhancer elements (Enh: H3K4me1, H3K9ac, and H3K27ac). ChromHMM was used to determine chromatin features using Encode data from E15 mouse forebrains (ChIP-seq: H3K4me3, H3K4me2, H3K4me1, H3K36me3, H3K9ac, H3K27ac, H3K27me3, and H3K9me3). (G,H) Representative genome browser tracks showing changes in accessibility peaks in KO and Het forebrains at regulators of NSPC identity and neural differentiation.