Figure 7.
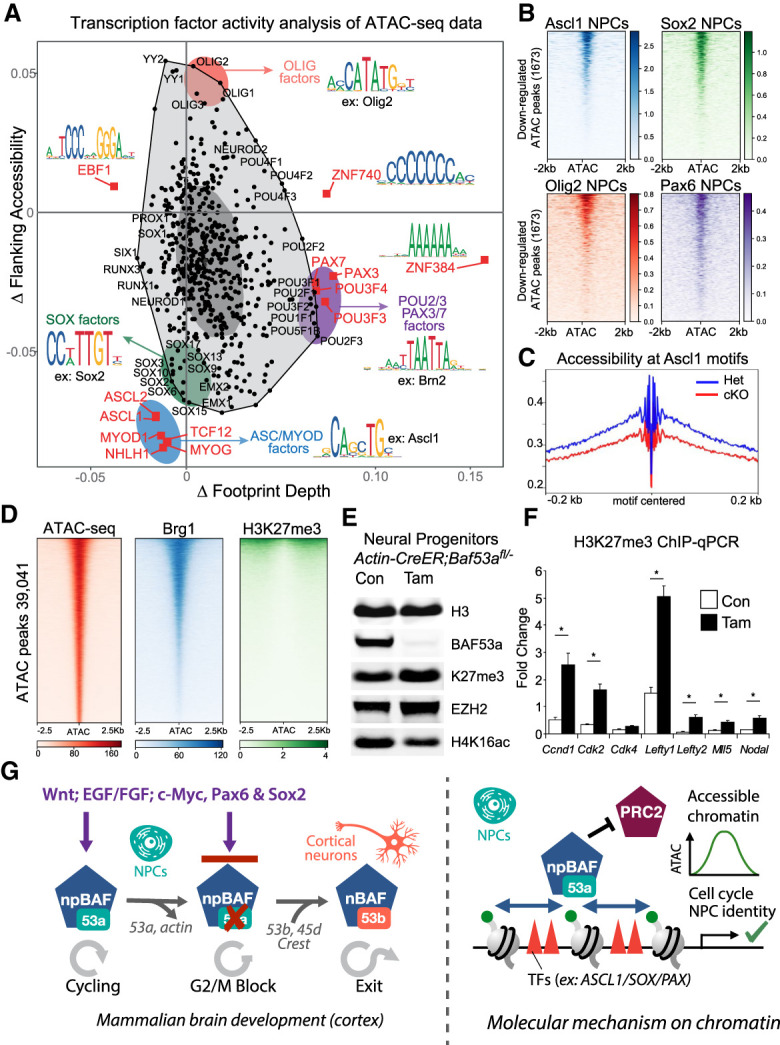
BAF53a regulates accessibility for neurogenesis TF activity through opposition to Polycomb repressive complex 2. (A) BaGFoot analysis of TF activity at BAF53a-dependent ATAC peaks. This analysis determines both changes in flanking accessibility, as well as changes in TF footprints. ASC, SOX, BRN, and PAX families of TFs displayed reduced activity in BAF53a mutant brains. TF binding motifs for a given member of each family are displayed as examples. (B) Analysis of neural TF binding at ATAC-seq peaks down-regulated in BA53a mutant (cKO) forebrains. We used published ChIP-seq data sets from mouse NPCs for ASCL1 (Wapinski et al. 2013), SOX2 (Mistri et al. 2015), OLIG2 (Nishi et al. 2015), and PAX6 (Sun et al. 2015) to show binding of these different TFs at BAF53a-dependent accessibility peaks. (C) Characterization of ATAC peaks at ASCL1 motifs in BAF53a mutant (cKO) and control (Het) forebrains at E15.5. Chromatin accessibility is reduced at ASCL1 motifs in mutant brains. (D) Analysis of ChIP-seq and ATAC-seq data sets to characterize BRG1 and H3K27me3 levels across accessibility peaks in wild-type mouse forebrains. The majority of accessible regions are bound by BRG1, whereas very few accessible regions are marked with H3K27me3. (E) Western blot analysis of H3K27me3 levels in Actin-CreER;Baf53afl/− NSPCs treated with tamoxifen for 72 h. (F) ChIP-qPCR analysis of H3K27me3 levels at cell cycle regulator genes, stem/progenitor cell maintenance genes, and Hox cluster genes. The levels of the H3K27me3 repressive marks are increased at cell cycle genes following Baf53a deletion in Actin-CreER;Baf53afl/− NSPCs. n = 3. Error bars represent SEM. (*) P < 0.05. (G) During cortical development, the switch from npBAF to nBAF chromatin regulators enables cell cycle exit and ensures faithful initiation and execution of postmitotic neural differentiation programs in the cortex. (Left panel) Continued expression of BAF53a-containing npBAF complexes in NSPCs promotes proliferation downstream from multiple cues (e.g., WNT and EGF/bFGF) by opposing the actions of PRC2, generating chromatin accessibility for cell type-specific TFs to drive expression of cell cycle and NSPC identity genes. When the npBAF subunit BAF53a is deleted, NSPCs can no longer respond to proproliferative signals and await expression of BAF53b and the other nBAF subunits to progress through neural differentiation programs. (Right panel) npBAF complexes regulate chromatin accessibility at cell cycle genes and NPC identity genes through opposition to repressive Polycomb complexes. (Right panel) The chromatin accessibility generated by npBAF remodeling complexes promotes the binding activity of neural identity TFs, including the pioneer TFs ASCL1 and SOX2.
