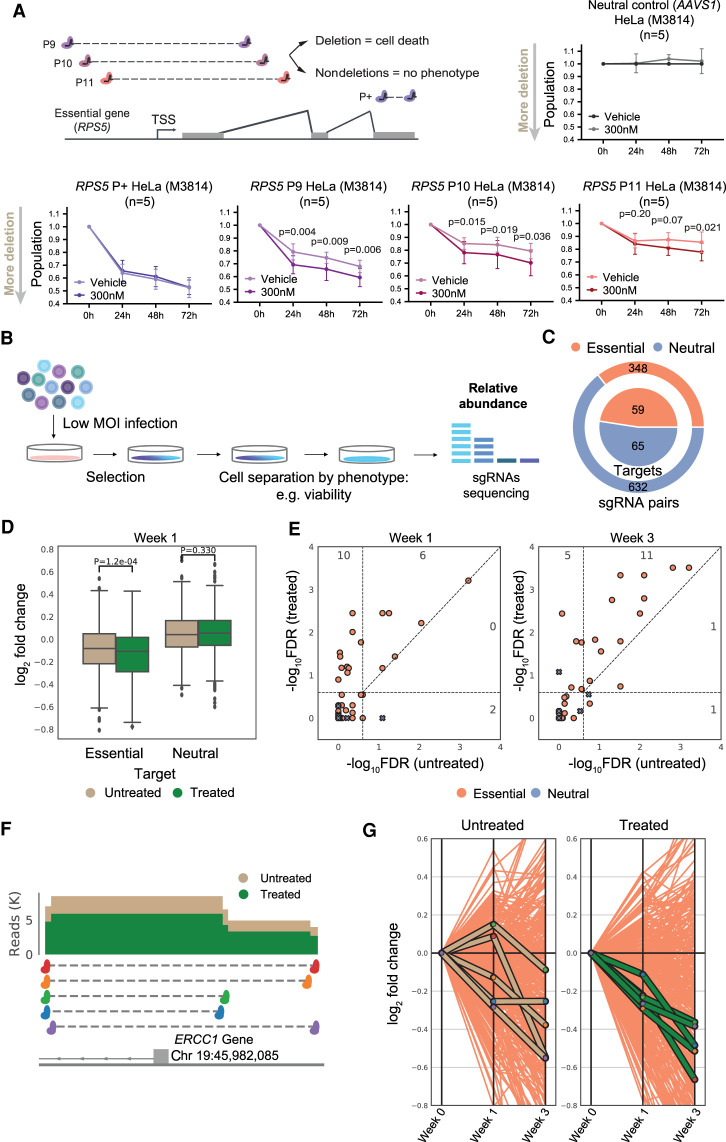
Figure 4.
Applying DNA-PKcs inhibition to pooled CRISPR-del screens. (A) Testing the functional benefit of DNA-PKcs inhibition. (Left) Experimental setup, with pgRNAs targeting the transcription start site (TSS) of the essential gene RPS5. A positive control sgRNA pair (P+) target the gene open reading frame and thus cause loss of function independent of deletion. (Right) Viability assays with pgRNAs targeting the nonessential AAVS1 locus. (Bottom) Viability assays after RPS5 TSS deletion (mean, standard deviation, one-tailed paired t-test). (B) Design of a typical pooled high-throughput screen to identify essential genes. (C) Composition of pgRNA library targeting both essential genes’ TSS and nonessential neutral loci. (D) Log2 fold-change (LFC) in abundance of indicated pgRNAs compared with day 0. Significance calculated by two-tailed t-test. (E) Hits reported by MAGeCK at two timepoints. Negative log10 false-discovery rate (−log10FDR) for treated and untreated samples are indicated in the y- and x-axis, respectively. Each point represents a target. A hit is called a true positive (TP) if it is targeting an essential gene and has an FDR <0.25. Points above the diagonal indicate hits with a lower FDR (higher −log10FDR) in the treated sample. Numbers in plot reflect TPs in each combination of treated/untreated cells. (F) pgRNAs targeting ERCC1 TSS: read coverage for untreated and treated samples at 1 wk. (G) Fold-change variation for individual ERCC1 pgRNAs across timepoints: x-axis, timepoint (0, 1, and 3 wk); y-axis, LFC in abundance. Orange lines represent all pgRNAs targeting essential genes.