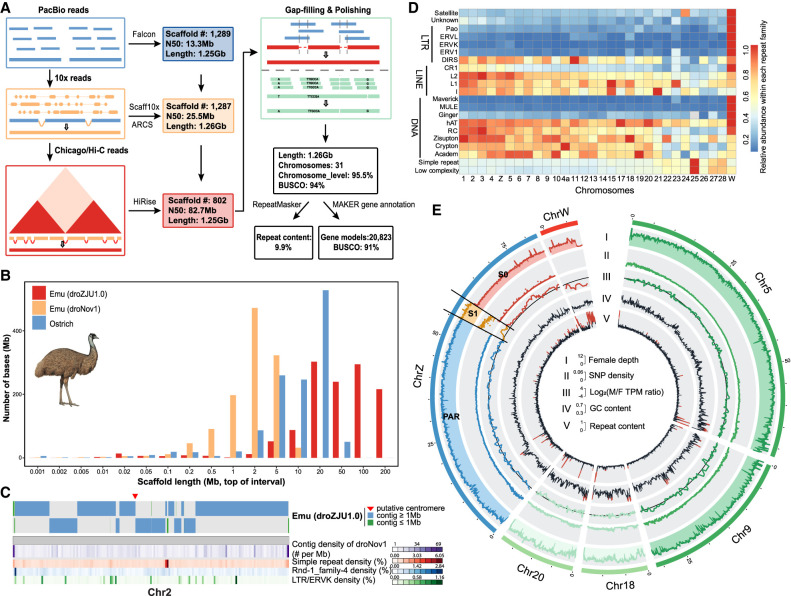
Figure 1.
Genome assembly of a female emu. (A) The PacBio long reads were first used to generate contigs, then used various linkage data of 10x linked reads, Chicago, and Hi-C reads to connect the contigs into chromosomal sequences, and finally polished the assembly with corrected PacBio long reads and Illumina reads. (B) Comparison of scaffold length distribution between droZJU1.0 and droNov1 emu assemblies and ostrich assembly, the latter two of which were generated by Illumina reads. (C) An example showing that the new droZJU1.0 assembly is more continuous than droNov1. In almost all the cases, one droZJU1.0 contig corresponds to multiple droNov1 contigs, and regions of high repeat content (e.g., LTR/ERVK) coincide with the breakpoints between contigs. (D) The abundance of each repeat family was normalized to a range from 0 and 1 for each chromosome, respectively. (E) The genomic landscape of emu chromosomes. We showed two macrochromosomes (Chr 5 and Chr 9), two microchromosomes (Chr 18 and Chr 20), and the Z/W sex chromosomes for their genomic compositions. (I) PAR (blue), autosomes (green), S1 (orange) show a twofold higher female read coverage than the S0 and Chr W (red). (II) S1 shows a higher female SNP density than any other genomic regions. (III) S0 shows a male-biased expression pattern. (IV) Microchromosomes have a higher GC content than macrochromosomes. (V) Microchromosomes have a lower repeat content than macrochromosomes.