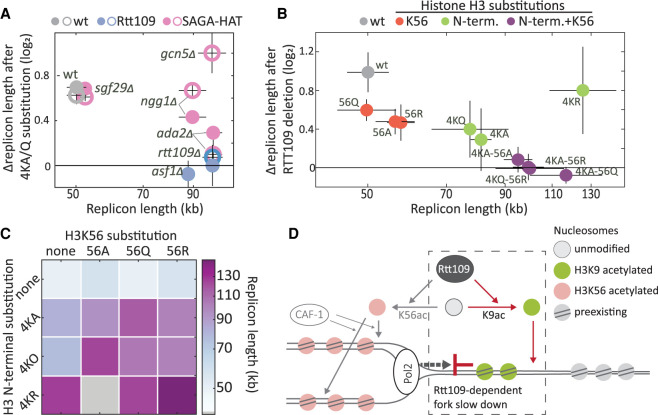
Figure 5.
Rtt109 acts via H3 N-terminal acetylation to reduce fork velocity. (A) Genetic interaction between histone N-terminal acetylation and Rtt109 or SAGA/SLIK complexes. Relative replicon length change (log2) after an N-terminal substitution (4KQ, open circle; 4KA, closed circle) in wt (gray), RTT109-deleted (blue), or SAGA-HAT-core subunit-deleted (pink) cells is plotted against their replicon length (x-axis in log scale). (B) Genetic interactions between RTT109 and histone acetylation during DNA replication. For each lysine H3 N-terminal and K56 substitution (naming as in Figs. 3A, 4A), and their combinations, the relative replicon length (log2) after RTT109 deletion is plotted against its replicon length without RTT109 deletion. Gray indicates wt; red, H3K56 substitutions; green, N-terminal substitutions; purple, combined H3K56 and N-terminal substitutions; x-axis in log scale. (C) Synergistic effect of H3K56 and H3 N-terminal substitutions on DNA replication dynamics. The replicon lengths of mutants with combinations of H3K56 (columns) and H3 N-terminal 4K (rows) substitutions are shown. Colored squares indicate the replicon length after combining N-terminal mutation; gray squares indicate missing data for 4KR + K56A. (D) A working model: Histone H3 N-terminal acetylation by Rtt109 promotes nucleosome deposition in front of the fork and thus slows DNA replication.