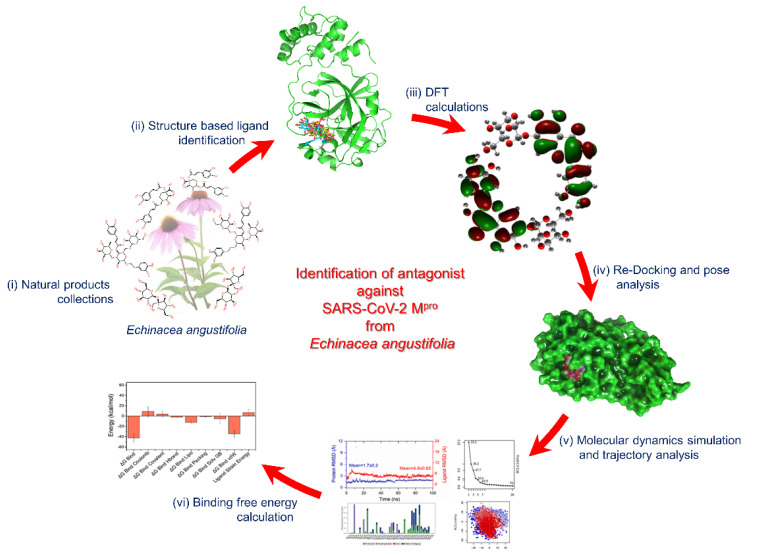
Figure 1.
Computational assessment of natural products in E. angustifolia against SARS-CoV-2 Mpro. Herein, (i) 3D structures of natural products reported in E. angustifolia were retrieved from the PubChem database, (ii) these natural products were then screened into the active region of SARS-CoV-2 Mpro by XP docking, (iii) natural products with highest negative docking energy were collected and treated by DFT method for respective geometry optimization to calculate other molecular properties, (iv) these optimized geometries of natural products were re-docked with SARS-CoV-2 Mpro and studied for pose binding and molecular contacts formation, (v) best docked poses were further studied for stability and protein ligand contact formation as function of 100 ns interval, and (vi) frames were extracted from respective MD trajectories and used in binding free energy calculations.