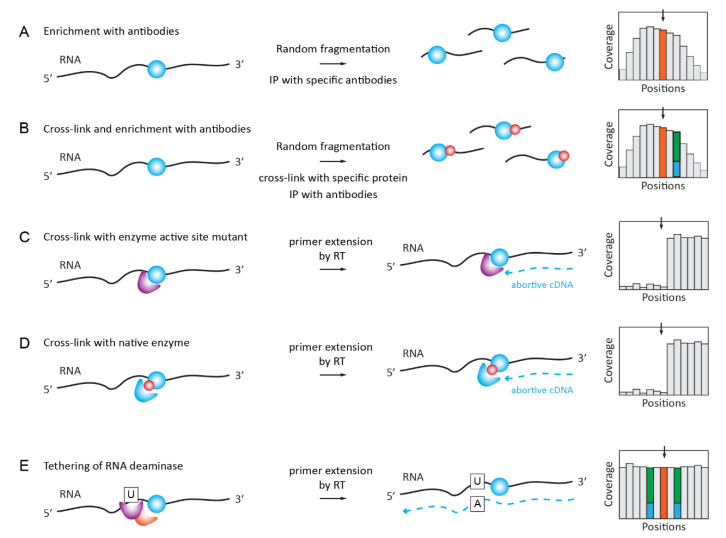
Figure 3.
Detection of RNA modifications by immunoaffinity and other affinity-related methods. (A): A classical Me-RIP approach first applied to m6A and further extended also to non-methylated modified nucleotides. RNA is fragmented and the modification-containing fraction is enriched by immunoprecipitation. The signal of Me-RIP is somehow asymmetric, and a modified nucleotide may be displaced from the peak summit. Panel (B): immunoprecipitation-based individual-nucleotide resolution UV crosslinking and immunoprecipitation/Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (iCLIP)/PAR-CLIP approaches using the covalent cross-linking of the specific antibody to RNA in the proximity of RNA modification. RT signature (shown in blue/green and resulting from RNA/protein cross-link may be visible in proximity to the modified nucleotide. Panel (C): A covalent cross-linking with the modified residue when the enzyme active site mutant is used. Described for m5C and m5U methylating enzymes, which use a two-step reaction mechanism. Panel (D): cross-linking with the native enzyme may be achieved by the incorporation of non-natural nucleotides in RNA in vitro or in vivo (metabolic labeling). Panel (E): Reader protein for RNA modification is fused with the RNA deaminase domain (APOBEC) and, thus, tethered to the modified RNA residue (shown in orange), inducing partial C→U deamination in close proximity to RNA modification. These partial deamination signatures are visible in the sequencing profiles (shown in green/blue).