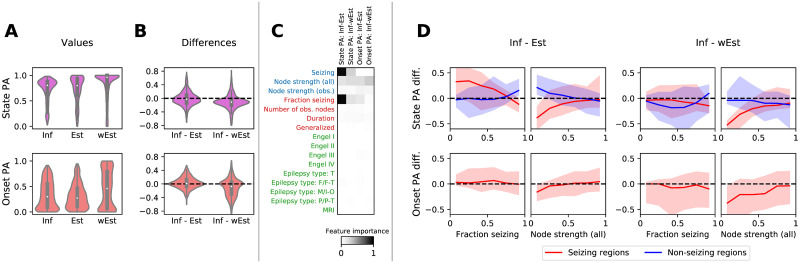
Fig 6. Results of the leave-one-out (LOO) validation.
(A) Distributions of the state prediction accuracies (PA), top, and onset prediction accuracies, bottom, calculated with the inference (Inf) method and the unweighted estimate (Est) and weighted estimate (wEst). In the upper panel, each data point (n = 2863) corresponds to one observed region in one seizure (nseizures = 137) of one patient (npatients = 44) for which LOO analysis was successfully performed. In the lower panel, only the seizing regions are considered (n = 1679). (B) Pairwise differences between the accuracies obtained by the inference and the two estimates. In both A and B, the violin plots show a kernel density estimate of a given variable. The inner boxplots show the median (white dot), interquartile range (IQR, gray bar) and adjacent values (upper/lower quartile +/- 1.5 IQR, gray line). (C) Feature importances for predicting the differences in B. On vertical axis, features on region level (blue), seizure level (red), and patient level (green) are shown. Horizontal axis contains the target variables, that is the four prediction accuracies differences. Higher values of feature importance indicate a stronger dependency of the target variable on the feature. Three features are identified as most relevant: seizing/non-seizing state of a region, node strength in a network, and a fraction of seizing region in a seizure. (D) The partial dependency plots of the PA differences on features identified by the feature importance analysis in C. These are the fraction of seizing nodes, and the node strength, both for seizing and non-seizing regions. Full line shows the median, and filled area represents the 10 to 90 percentile range.