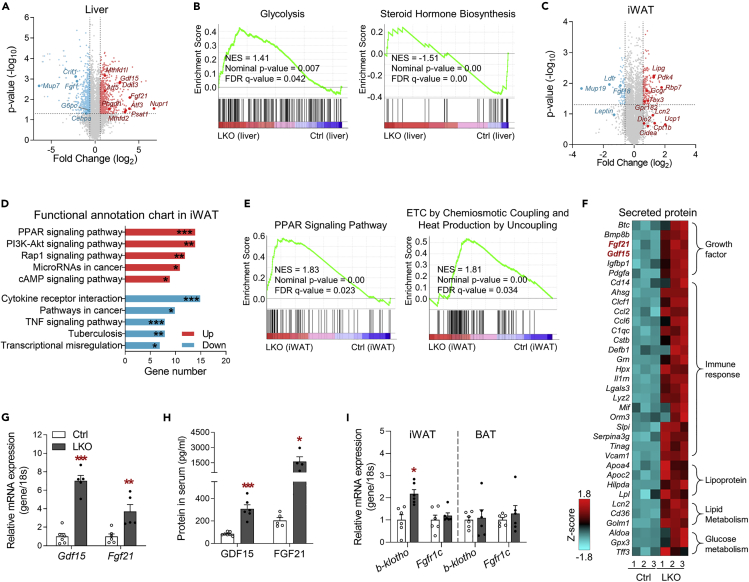
Figure 4.
Liver-specific Crif1 deficiency increases the production of metabokines in liver
(A) Volcano plot showing the DEGs in the liver of LKO mice. The colored dots indicate the DEGs with ≥ ±1.5-fold difference from Ctrl mice. The red and blue dots indicate the upregulated and downregulated transcripts, respectively.
(B) Gene set enrichment analysis (GSEA) using DEGs in the liver of Ctrl and LKO mice.
(C and D) Volcano plot showing the DEGs (C) and top-ranked functional annotation chart (D) in the iWAT of LKO mice. Functional annotation was categorized using the KEGG pathway in DAVID (ver.6.8), and the results are ordered according to gene number.
(E) GSEA showing the upregulated gene set in the iWAT of LKO mice.
(F) Heatmap of the DEGs classified as “Secreted proteins” in the cellular compartment category (≥2-fold difference, p < 0.05 versus Ctrl mice). The genes in red were the two top-ranked transcripts, Fgf21 and Gdf15 in the liver of LKO mice.
(G) Quantitative PCR analysis of Gdf15 and Fgf21 mRNA expression in Ctrl and LKO livers (n = 5–6 biological replicates from four independent experiments).
(H) Serum GDF15 (n = 6–7 biological replicates) and FGF21 (n = 4–5 biological replicates) in Ctrl and LKO mice. Three independent experiments were performed.
(I) Quantitative PCR analysis of Klb and Fgfr1c mRNA expression in adipose tissue of mice (n = 6 biological replicates).
The mice were studied at 8–9 weeks of age and fed a chow diet. Data in (G–I) are mean ± SEM. Statistical analyses were performed using Student's t test in (G–I) (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus Ctrl). The data in (D) were analyzed using a modified Fisher's exact p value (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Un.d, undetectable. See also Figure S4.