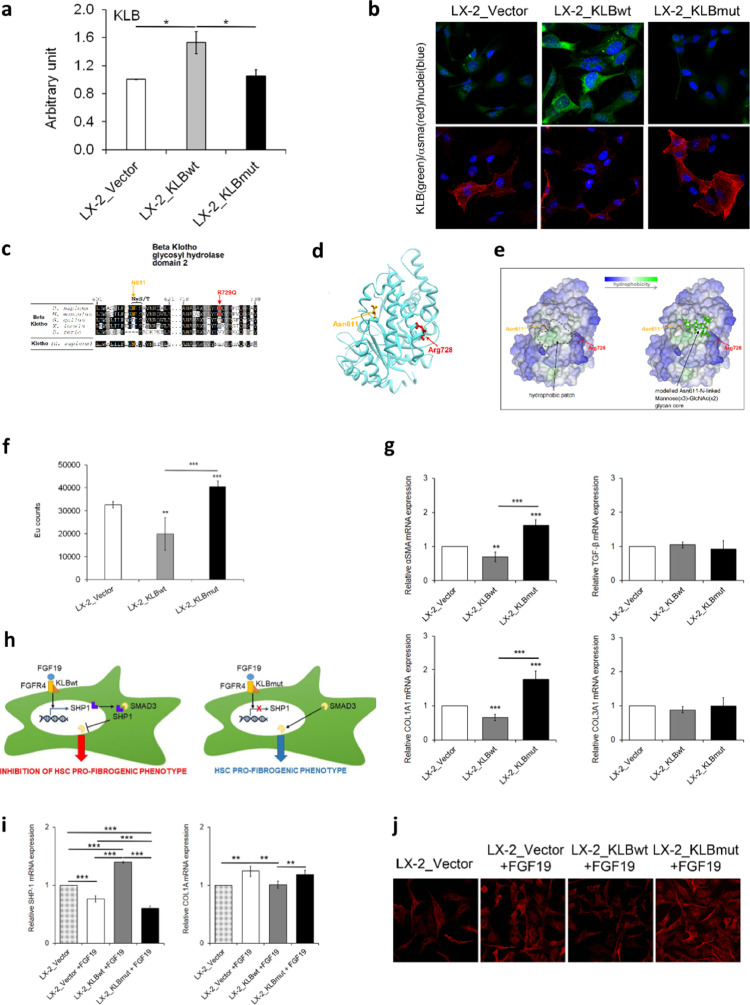
Fig. 5.
Analysis of the effect of wild-type and mutant KLB overexpression in LX-2 cells. In this experimental setting LX-2 cells were transiently transfected with pCMV6 plasmid (LX-2_Vector), KLB wild-type plasmid (LX-2_KLBwt), or KLB mutant plasmid (LX-2_KLBmut), and all analyses were performed after 48 h. Relative expression of KLB protein levels was determined by densitometric analysis of Western blots. The intensity of the bands was analyzed by ImageJ software (a). Representative immunofluorescence images of cellular distribution of KLB (green) and αSMA (red). Nuclei are reported in blue. Magnification 60X (b). Multiple sequence alignment around the site of the Arg728Gln variant among orthologues of KLB and the human Klotho paralogue. Consensus and similar residues are respectively in black and gray background. The consensus residues NxS/T (where “x” can be any residue except proline) for the N-glycosylation of Asn611 are indicated (c). The structure of the KLB GH2 domain isolated from the Protein Data Bank entry 5VAN and the modelled to show the position of Arg728, the N-glycosylable Asn611 (d). Same KLB GH2 domain structure as in D represented as molecular surface colored by residue hydrophobicity, highlighting the surface-exposed protein hydrophobic patch (left) and showing a modelled mannose(x3)-GlcNAc(x2) glycan N-linked to Asn611 (e). Cell proliferation was evaluated by a BrdU incorporation kit and expressed as Eu counts (f). mRNA levels of αSMA, TGF-β, COL1A1 and COL3A1 were evaluated by qRT-PCR (g). Hypothesis of mechanism by which KLB exert the inhibition of pro-fibrogenic genes after LX-2 cells stimulation with FGF19 (h). mRNA levels of SHP-1 and COL1A1 were evaluated by qRT-PCR (i). Representative immunofluorescence images of cellular distribution of SMAD3 (red). Magnification 40X (j). Quantitative data are the mean ± SD of 2 independent experiments repeated at least in triplicate. Data were analyzed by two-way ANOVA with Bonferroni post hoc test for multiple comparisons. Adjusted *p<0.05; **p<0.01; ***p<0.001.