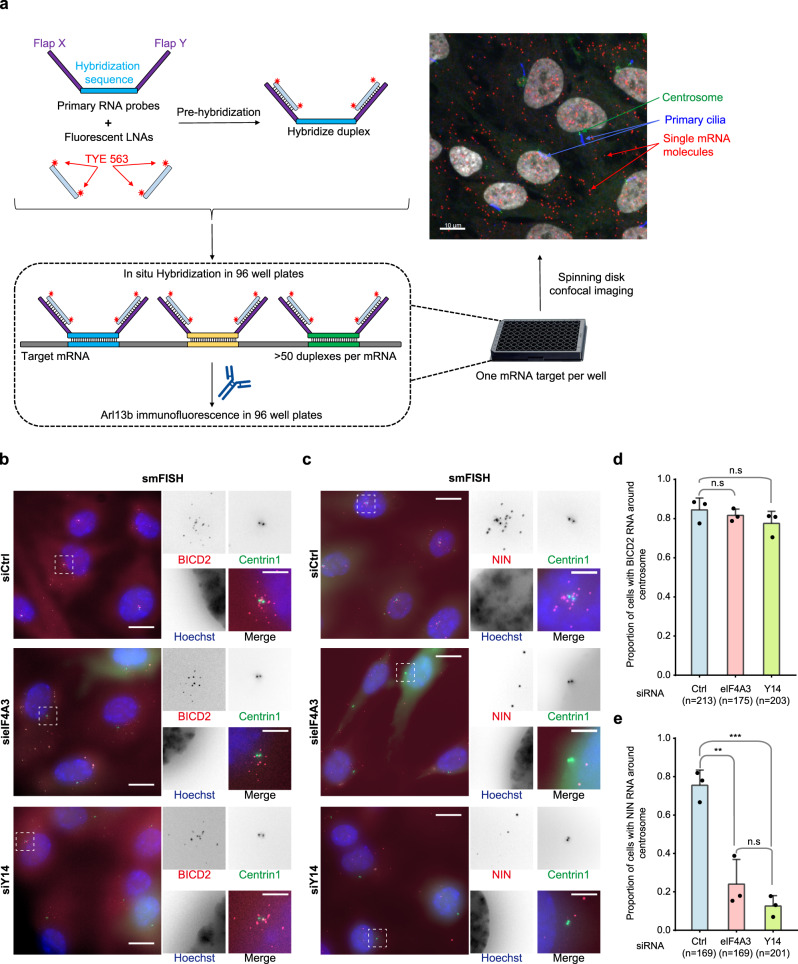
Fig. 5. EJC is required for centrosomal localization of NIN mRNA.
Summary of the high-throughput smiFISH pipeline (a). Top left: primary RNA probes contain a hybridization sequence that is complementary to the target mRNA flanked by two overhangs named Flap X and Y. Each Flap was annealed to a locked nucleic acid (LNA) oligo labeled with two TYE 563 molecules in a pre-hybridization step. Bottom: Duplexes were then hybridized to the mRNA of interest followed by immunofluorescence against Arl13b to label primary cilia in 96-well plates. Plates were finally imaged with a spinning disk confocal microscope. Top right: A micrograph showing a typical field of view from the screen. Red dots correspond to a single mRNA molecule. Scale bar represents 10 μm. Quiescent RPE1 cells stably expressing centrin1-GFP were stained by probes against BICD2 mRNA (b) or NIN mRNA (c) after knock-down of either eIF4A3 or Y14. Nuclei were stained by Hoechst. Images are resulted from maximum intensity projections of 14 z-stacks acquired at every 0.5 μm. Right panels show enlarged images of the white dashed square in the left panels. Scale bars in the left panels are 10 μm, and scale bars in right panels are 3 μm (b, c). Proportion of cells displaying centrosomal BICD2 (d) or NIN (e) RNA pattern was depicted. Columns and bars depict mean ± SD of three independent experiments. n.s P > 0.05, **P ≤ 0.01, and ***P ≤ 0.001, two-tailed t-test. The number of cells analyzed in three independent experiments is provided (d, e). Source data are provided as a Source Data file.