Abstract
Three coronaviruses (CoVs) have threatened the world population by causing outbreaks in the last two decades. In late 2019, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) emerged and caused the coronaviruses to disease 2019 (COVID-19), leading to the ongoing global outbreak. The other pandemic coronaviruses, SARS-CoV and Middle East respiratory syndrome CoV (MERS-CoV), share a considerable level of similarities at genomic and protein levels. However, the differences between them lead to distinct behaviors. These differences result from the accumulation of mutations in the sequence and structure of spike (S) glycoprotein, which plays an essential role in coronavirus infection, pathogenicity, transmission, and evolution. In this review, we brought together many studies narrating a sequence of events and highlighting the differences among S proteins from SARS-CoV, MERS-CoV, and SARS-CoV-2. It was performed here, analysis of S protein sequences and structures from the three pandemic coronaviruses pointing out the mutations among them and what they come through. Additionally, we investigated the receptor-binding domain (RBD) from all S proteins explaining the mutation and biological importance of all of them. Finally, we discuss the mutation in the S protein from several new isolates of SARS-CoV-2, reporting their difference and importance. This review brings into detail how the variations in S protein that make SARS-CoV-2 more aggressive than its relatives coronaviruses and other differences between coronaviruses.
Keywords: Coronaviruses, Spike proteins, SARS-CoV-2, SARS-CoV, MERS-CoV, RBD, Mutations
1. Introduction
Coronaviruses (CoVs) belong to a family of positive-sense, single-stranded, RNA viruses that are lipid-enveloped [1]. The classification by virologists classified coronaviruses into four genera as alpha, beta, gamma, and delta. The most famous are the ⍺- and β-coronaviruses, given their ability to pass through the animal-human barriers, thus becoming clinically relevant to humans [2]. Nowadays, virologists reported seven coronaviruses able to infect humans and named as human coronaviruses (hCoVs). Of these, human coronavirus OC43 (hCoV-OC43), Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), Human coronavirus HKU1 (hCoV-HKU1), MERS-CoV are classified into the beta-genera, and human coronavirus NL-63 (hCoV-NL63) and Human coronavirus 229E (hCoV-229E) into the α-genera [1,[3], [4], [5], [6]].
The hCoVs hCoV-HKU1, hCoV-OC43, hCoV-NL63, and hCoV-229E are not that harmful to humans as their infection results in non-symptomatic infection or, worse case, mild respiratory and less common gastrointestinal infection. Today, 5–30% of common cold cases are attributed to these hCoVs. Therefore, humans did not give the necessary attention to the hCoVs. However, in the last two decades, three outbreaks caused by hCoVs made humans pay more attention to them. The ongoing outbreak showed the human population how devastating hCoVs could be and driven many researchers worldwide to find either a vaccine or a treatment [[1], [2], [3], [4], [5], [6]].
In December 2019, China warned the World Health Organization (WHO) about pneumonia caused by a new virus in Wuhan [[7], [8], [9]]. Later, the new virus was grouped into the coronavirus family and named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). SARS-CoV-2 spreads quickly worldwide, and in a few months, more precisely in March 2020, the situation became worse and established a pandemic state [[9], [10], [11]]. The pandemic situation was followed by lockdown strategies adopted by the entire world. Despite that, the health care systems around the world went through high pressure, followed by the shutting down of the economic situation in many countries [7].
Unlike the 2002–2003 SARS-CoV-1 outbreak, SARS-CoV-2 infection reached all continents quickly, proving to be more contagious. Comparing both viruses, the infection caused by SARS-CoV-2 is recognized by a broader clinical spectrum that comes from asymptomatic infection to severe viral pneumonia with respiratory failure and death [8,12,13]. In contrast to SARS-CoV and MERS-CoV, many SARS-CoV-2-infected patients developed low-titer of neutralizing antibody, leading them to suffer with prolonged severe symptoms and illness, suggesting a more effective SARS-CoV-2 immune surveillance evasion than SARS-CoV and MERS-CoV [[14], [15], [16], [17], [18]].
The ongoing outbreak caused by SARS-CoV-2 has taken many lives, threatened thousands more, and destroyed entire families worldwide. Indeed, SARS-CoV-2 (Coronavirus Disease 2019) is by far more transmissible than SARS-CoV-1 and MERS-CoV but is less lethal than they are (Table 1 ). However, SARS-CoV-2 higher transmissibility has resulted in 113,299,920 million of infected people with 2,512,823 million of death as of 25 February 2021, by far a larger number compared to other outbreaks [19,20].
Table 1.
Human coronaviruses (hCoV) related-symptoms and their case fatality rates.
| hCoV | Case fatality rate | Symptoms | Refs |
|---|---|---|---|
| SARS-CoV | 9.6% | Fever Myalgia Headache Malaise Chills Nonproductive cough Dyspnea Respiratory distress Diarrhea |
[42,56,113] |
| MERS -CoV | 35.5% | Fever Cough Chills Sore throat Myalgia Arthralgia Dyspnea Pneumonia Diarrhea and vomiting Acute renal impairment |
[98,113] |
| SARS-CoV-2 | 6.8% | Fever Cough Chills Myalgia Arthralgia Dyspnea Pneumonia Diarrhea and vomiting |
[9,111,113] |
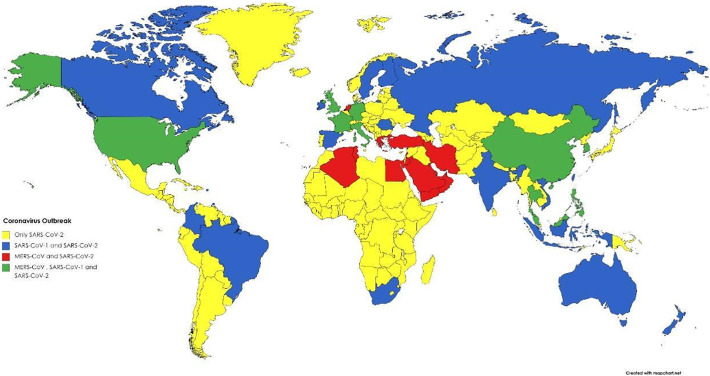
The coronaviruses outbreak presented different spreads worldwide. Starting in November 2002 in Foshan-China, until March 2003, the SARS-CoV-1 outbreak has spread all over 29 countries (Fig. 1 ) [21,22]. The WHO status for SARS-CoV-1 today is controlled [21]. The MERS-CoV outbreak started in April 2012 in Zarqa-Jordan, and by September 2012, spread to 27 countries (Fig. 1) [[22], [23], [24], [25]]. The status of MERS-CoV by WHO is sporadic continuous [24]. For SARS-CoV-2, as we know, the status of the outbreak is ongoing and has already spread to 213 countries (Fig. 1) [26].
Fig. 1.
Worldwide view of Coronaviruses outbreak hotspots: spread locations of SARS-CoV 1, SARS-CoV 2, and MERS-CoV around the globe. Yellow represents dissemination of only SARS-CoV-2, blue represents dissemination of both SARS-CoV-1 and SARS-CoV-2, red represents dissemination of MERS-CoV and SARS-CoV-2, and green represents dissemination of the three coronaviruses. Created with BioRender.com.
The higher transmissibility of the SARS-CoV-2 is probable due to the higher number of accumulated positive mutations in the spike glycoprotein (S), which led this protein to be 20 times more effective in recognized human receptors than the spike from SARS-CoV-1 and MERS-CoV [27,28]. Based on that, the review is focused on the discussion and tracking the mutations in the S protein in three recent coronaviruses that posed outbreaks of SARS-CoV-1, MERS-CoV, and SARS-CoV-2. Additionally, we intend to understand the contribution of the S protein to the transmissibility of SARS-CoV-2 compared to SARS-CoV-1 and MERS-CoV.
2. Coronaviruses taxonomy and linking to bats
The coronaviruses are one of the two genera belonging to the Coronaviridae family. The other one is the toroviruses [[29], [30], [31]], both included in the Nidovirales order. Coronaviruses are highly disseminated among mammals in general, causing mild infections such as cold and gastrointestinal. However, in some cases, they might cause severe respiratory infection. In the past, coronavirus was known the most by an infection caused in animals such as bronchitis virus (IBV) in chickens and pigs and Feline infectious peritonitis (FIP) in cats [[7], [8], [9], [10], [11],32].
Nowadays, at least 60 coronaviruses have been isolated and described, mainly from bats (BtCoVs). Most of the coronaviruses identified belong to the betacoronavirus group. It has been accepted that the power to flight of bats associated with the ability to migrate to regions far from the original spot has imposed a strong selective pressure for the coexistence with viruses [[33], [34], [35], [36]]. Additionally, bats' immune system is known as permissive, which means that bats can act as a reservoir to coronaviruses without developing the disease [[37], [38], [39], [40]]. Bats, at the same time, could act as a carrier to 10 up to 17 α-coronaviruses and 7 up to 12 β-coronaviruses with the potential to jump to humans passing interspecies barrier causing disease [41]. To corroborate that, long-term studies by genome sequencing revealed nucleotide similarity between coronaviruses found in humans and bats using the same cell entry, the Angiotensin-Converting Enzyme 2 receptor (ACE2) [38]. These studies [[33], [34], [35], [36], [37], [38], [39], [40], [41]] helped to track back the hCoVs to bats. For instance, it is hypothesized that the MERS-CoV evolved from a progenitor hosted in bats to infect dromedary and camels with the ability to directly infect humans [6].
3. The pandemic coronaviruses
3.1. SARS-CoV
In 2002, SARS-CoV-1 emerged in Guangdong Province, China, that spread over five continents 29 countries, leading to 8098 cases and 774 deaths in nearly September 2003 [[42], [43], [44]]. The SARS-CoV was the first threat imposed by hCoVs to humankind that outbreaks around the world. During the outbreak, the estimated case fatality rate (CRF) was 9.6%, and the human-to-human transmission of SARS-CoV was confirmed. On 30 January, WHO, warned by China scientific council, declared the SARS-CoV outbreak as an international public health emergency [21]. The first SARS-CoV cases occurred in employees who worked in a restaurant that handled wild mammals served as exotic food. Studies found that Chinese horseshoe bats had sequences of SARS-related CoVs and hosted a virus that shares similarities with SARS-CoV. Based on that, the origin of SARS-CoV is believed to have occurred in Chinese horseshoe bats [44,45].
The first suspected source of SARS-CoV was civets, a small mammal, due to the detection of the virus in those animals. Nonetheless, these animals are only transient hosts and found no evidence in wild civets [46]. Meanwhile, the evidence points to bats as hosts for SARS-CoV since they are permissive to SARS-CoV-like viruses [46].
3.1.1. Origin and evolution of SARS-CoV-1
SARS-CoV belongs to β-coronaviruses (Fig. S1) of the Coronaviridae family and is involved in zoonotic transmission and spread among humans through close contact [47]. It was later reported the first patient with SARS-CoV had prior contact with animals before developing the symptoms. The causative agent of SARS was later found in palm civets. However, strains isolated from handled market civets were transmitted from other animals. This suggests civets only as an intermediate host. Based on that, the hypothesis that the bat was the natural host of SARS-CoV came back to the spotlight [48,49].
Hu et al. [50] provided new information suggesting that the horseshoe bat (Rhinolophus sinicus) is the natural host of SARS-CoV. The authors reached that conclusion after the isolation of SARS-like CoVs that was homologous to SARS-CoV. Moreover, evidence shows the possible origin of SARS-CoV was based on the recombination of different SARS-like coronavirus in bats since some potential recombination sites were identified around the S gene [50,51]. Rest and Mindell [52] tried to elucidate the phylogenetic origin of SARS-CoV by comparing RNA dependent RNA polymerase (RdRp) sequences between SARS-CoV and other coronaviruses. The results showed that the SARS-CoV sequence is a recombinant virus. Recently, phylogenetic analyses showed a high similarity (80%) between SARS-CoV and SARS-CoV-2 [53]. Regarding the S gene, the similarity between SARS–CoV-2, and SARS-CoV is about 76% [47,54].
Coronaviruses often undergo mutations and genetic recombination, as they have error-prone RdRP, which results in a bigger diversity, adaptive evolution, and capacity to cause disease. Previous studies have shown that SARS-CoV mutated over the 2002 and 2004 epidemic to better bind to its cellular receptor (ACE2) and replication in human cells, enhancing virulence [55]. The S1 subunit of S protein has a receptor-binding domain (RBD) involved in the direct interaction with the ACE2 receptor [47,[53], [54], [55]].
3.1.2. Pathogenicity
During the SARS-CoV epidemic in 2002, patients usually presented fever, myalgia, headache, malaise, chills, cough, dyspnea, and respiratory distress generally 5 to 7 days later, resulting in death. In some cases, the gastrointestinal tract, liver, kidney, and brain [56]. SARS-CoV infection of the lungs leads to diffuse alveolar damage, epithelial cell proliferation, and an increase in macrophages [57,58].
The high transmission efficiency of the SARS-CoV occurs because it binds to a target (ACE2) on cells that are abundantly expressed, including pneumocytes in the respiratory system. The virus enters and replicates in these cells. Thus, mature virions are then released to infect new target cells [47,52]. SARS-CoV-1 has an incubation period of ~5 days, and 95% of patients develop the disease within 13 days of exposure [46]. SARS-CoV was reported to cause the respiratory system and the gastrointestinal and other organ systems [56]. This is because the SARS-COV human receptor ACE2 is abundantly expressed in the lungs and small intestine [55,57,58].
SARS-CoV has unique pathogenesis because it causes upper and lower respiratory tract infections [55]. Common early symptoms are fever, chills, coughing, malaise, myalgia, headache, and less common symptoms, including diarrhea, vomiting, and nausea. In some cases, not common, SARS-CoV is associated with thrombotic complications and hematologic manifestations [42]. Abnormal chest X-rays are detected in roughly 60% of patients infected with SARS-CoV. It was reported that 20–30% of patients infected with SARS-COV require intensive care and mechanical ventilation [[59], [60], [61]]. In addition, at that time, it was noticed the SARS-CoV had developed high stability in aerosol and other surfaces, which had improved its transmissibility [62,63].
3.2. MERS-CoV
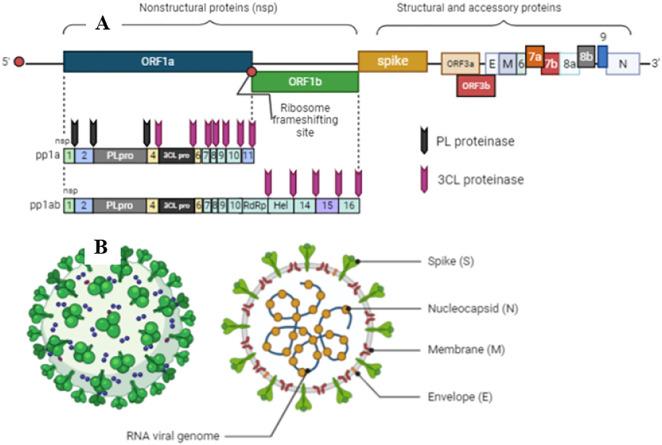
The MERS-CoV was primarily identified in Arabian Peninsula in 2012 as the major agent of a severe respiratory condition with a high case fatality rate (CFR) of ~35% (Table 1) [24]. Saudi Arabia was the first country to report MERS-CoV and the hotspot to its outbreak. MERS-CoV has officially 2279 laboratory-confirmed cases with 806 associated deaths in 27 countries (Fig. 1) [23,24,42,48,64,65]. Nowadays, there are yet reported cases of MERS-CoV infection. For example, WHO [24] has reported nine confirmed infection cases by MERS-CoV with five deaths from April to September 2020. It is proposed that during replicating the genome, the mutation rate of MERS-CoV is 4.81 × 10−4 substitutions per site per year [65,66]. MERS-CoV has the same genome size and produces the same proteins (Fig. 2 ) of SARS-CoV, which will be in-depth, discussed later [67].
Fig. 2.
Schematic diagram of coronaviruses genome and structural proteins of viral particles. A. Genome of coronaviruses produces non-structural proteins (nsp), such as RNA-dependent RNA polymerase involved in genome replication and two proteases involved in polyprotein processing, and structural and accessory proteins involved in composition of viral particles. B. The Genomes of coronaviruses also produce four structural proteins with significant roles in transmission and pathogenesis: spike (S), envelope (E), membrane (M), and nucleocapsid (N). Created with BioRender.com.
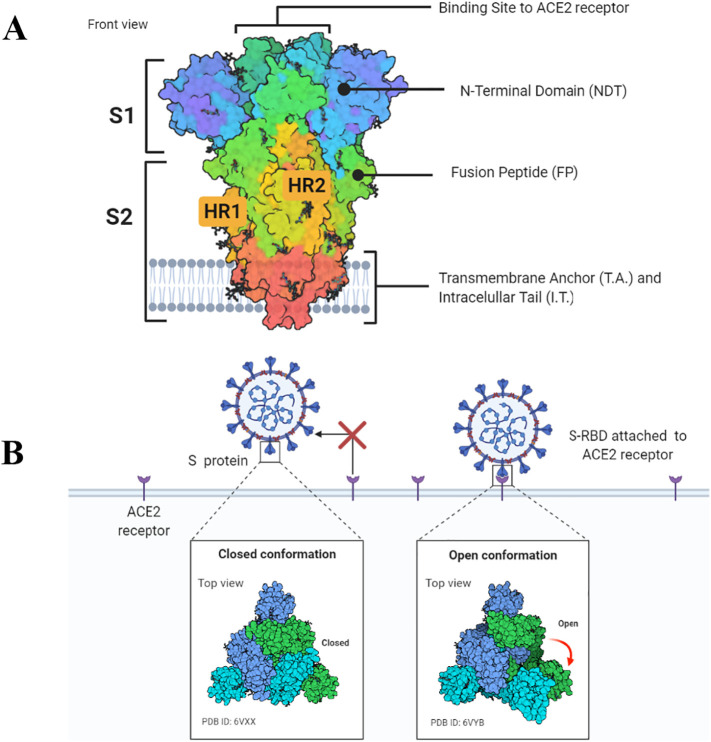
During the infection process, the S glycoprotein plays a central role in cell recognition, attachment, cell entry, and infection [27,28]. Structurally, the S protein is a trimetric protein with two subunits, S1 and S2, in each monomer (Fig. 3A). The S protein from MERS-CoV binds to the cellular receptor dipeptidyl peptidase 4 (DPP4/CD26) driven by S1. At this point, the MERS-CoV infection differs from that presented by SARS-CoV and SARS-CoV-2, which employ interaction with ACE2 receptor (Fig. 3B). SARS-CoV-2 is the only known CoV that could use both ACE2 and DPP4/CD26 receptors. The second step of infection is the same for the three pandemic coronaviruses, which involve the viral membrane and host membrane fusion and virus arrivals into the cytoplasm [64]. The replication process inside the cytoplasm will be later discussed in this review.
Fig. 3.
Schematic representation of the SARS-CoV-2 spike protein. A. Spike protein consists of the S1 and S2 units. In the S1 subunit, there is an extracellular N-terminal domain and a receptor-binding domain which play a role in the viral entrance into the cell through ACE2 receptor. In the S2 subunit, there is the fusion protein (FP), heptapeptide repeat sequence 1 and 2 (HR1 and HR2), as well as the transmembrane domain and a short C-terminal domain. B. Open or closed three dimensional conformation of S protein implicate in the binding of cell receptor ACE2. Created with BioRender.com.
3.2.1. Origin and evolution of MERS-CoV
Since the MERS-CoV outbreak, research worldwide has put together pieces of information to decipher the puzzle about MERS-CoV origin, pathogenicity, and transmissibility. The zoonotic event played a nontrivial part in MERS-CoV evolution and transmission [66]. Because a serosurvey study revealed that dromedary camels have antibodies against MERS-CoV, indicating that MERS-CoV has circulated in that area a long time before the outbreak. Harbor viruses that are closely genetically related to humans, the dromedary camels, constitute a source of pathogens that can be harmful to humans [[68], [69], [70], [71], [72]]. However, as to other coronaviruses, bats are believed to be the primary source of MERS-CoV origin and still works as a reservoir host of MERS-CoV. Genome sequence data revealed a similarity of 99.2–99.5% between bat and human MERS-CoV [23,48,68,69,71].
By analyzing 5030 fecal specimens from bats, Annan et al. [73] strongly suggested MERS-CoV likely comes from bats. Studies employing phylogenetic analysis showed two main MERS-CoV clades: A and B hosting camel and human MERS-CoV. MERS-CoV strains from humans from Jordan and Saudi Arabia belong to clade A. In contrast, clade B, divided into five groups (I to V), hosts MERS-CoV collected of humans from other regions and camels. The MERS-CoV from bats and hedgehogs forms a basal paraphyletic group to all camel and human MERS-CoV clades. This result suggests that, in addition to bats, hedgehog could also be the ancestor of camel and human MERS-CoV [36,66].
Additionally, it is known that recombination events can create new viral strains capable of infecting new hosts and evading immune responses from hosts. Overall, those mutations affect the S protein involved in both processes. There is evidence that recombination events on clade B groups, involving camel MERS-CoV and human MERS-CoV of different groups (I to V), produced different recombinant virus types (1 to 7) [66]. It was reported these recombination events might happen frequently and involving group V might happen broadly. Additionally, multiple recombination events indicate double infection, persistent infection, and superinfection during the transmission history [66].
The most recent common MERS-CoV ancestor was estimated as of March 2012, consistent with initial case detection and the beginning of the outbreak [74]. The genetic mechanisms underlying cross-species jumping remain poorly understood. However, it was reported that mutations on MERS-CoV S protein changed its surface charge, enhancing cross-species jumping and viral entry on cells through suboptimal DPP4 receptor [[75], [76], [77], [78]].
The higher number of positive selections in the S protein is completely comprehensive given the central role played by S protein in MERS-CoV infection [66]. Most of the positive selection sites were in the RBD of S protein. RBD has two portions, one binding region, and one core region. Both are crucial to the virus recognizing and entering host cells and it. It was observed at two sites of mutations in the binding region of RDB, suggesting these amino acid substitutions might improve MERS-CoV binding capability to bind to different host cells and thus facilitate its cross-species transmission [66]. Chen et al. [79] state that most of the recombination and sequence diversity is in the S gene. Those changes may affect the structural conformation of RBD and the interactions with cognate human receptors.
As discussed before, the receptor used by MERS-CoV in the cells is different from that employed by SARS-CoV and SARS-CoV-2. SARS-CoV and SARS-CoV-2 employ the ACE2 receptor, MERS-CoV picked DPP4 [80,81]. As SARS-CoVs, the S1 C-terminal (CTD) of MERS-CoV acts as RBD, with two domains: the core and receptor-binding motif (RBM) [79,[82], [83], [84]]. S1-CTD from MERS-CoV is structurally similar to that from SARS-CoVs, composed of a major β–sheet scaffold. However, while in SARS-CoVs, the RBM has many unordered structures forming loops, the MERS-CoV RDM mainly contains stranded β–sheets. It is postulated these structural differences were responsible for receptor-specificity between viruses [85].
3.2.2. Pathogenicity
The high similarities among the DPP4 from humans, camels, horses, and bats allow MERS-CoV to infect them. However, mutations in the DPP4 receptor from mice, hamsters, and ferrets prevent MERS-CoV infection [[86], [87], [88]]. MERS-CoV isolates sampled from humans and camels are highly similar to each other and use the human DPP4 receptor efficiently [89]. Studies revealed that these mutations change critical residues in mouse DPP4 compared to humans DPP4 affecting host specificity of MERS-CoV. Some of these residues mapped in mouse and hamsters are different from humans; two residues 288 and 330 in mouse DPP4, and five in 291, 295, 336, 341, and 346 in hamsters DPP4 lead to the incompatibility of DPP4 from mouse and hamster with MERS-CoV S proteins thus preventing the infection [89].
Interaction between the residues 330 of DPP4 with MERS-CoV RBD Y499 has been suggested as a key interaction [82,83]. Besides that, Peck et al. [87] observed that the residue 330 mutation knocks out an NXT glycosylation motif in mouse DPP4. These results show that glycosylation acts as a determinant of DPP4-mediated host range and inhibits infection by MERS-CoV. Other nonpermissive hosts (ferret, hamster, guinea pig) also have a nonconserved glycosylation site in the region of DPP4 that interacts with the MERS-RBD. Peck et al. [86] demonstrate that glycosylation is an essential barrier to MERS-CoV infection, yet other species-specific determinants are also responsible. Elegant experiments involving mutations of these residues from mice and hamsters to those correspondents in human DPP4 allow the MERS-CoV to infect mice and hamsters [[82], [83], [84],86,88,89]. As expected, S protein is a crucial determinant factor of host range MERS-CoV and alteration either on its structure or in its receptor dramatically affects virus success.
Overall, people infected and confirmed by a laboratory test for MERS-CoV were around 49 years old; 65% are males. The hospitalization time for patients to come back healthy was about 41 days [90,91]. MERS-CoV has a median incubation period of 5.2 days, reaching 12 days in normal people conditions, and a more extended period in patients immunocompromised and comorbidities [92,93]. The onset of symptoms to death 11.5 days. Even nowadays, the morbidity is still 36% (WHO, 2020), with more than 50% of patients showing viral accumulation on lungs revealed by radiography of the chest. Generally, the high viral loads on the lungs lead to acute respiratory distress syndrome [92,94,95].
The clinical manifestations of MERS-CoV could be in three forms. First, asymptomatic. Second, flu-like symptoms, cold, fever, cough, dyspnea, pneumonia, myalgia, diarrhea, vomiting, abdominal pain, chills or rigors, and malaise (Table 1). Third, acute progressive infection leading to acute respiratory distress syndrome, septic shock followed by multiorgan failure, and death [[96], [97], [98], [99], [100], [101]]. Nearly 50% of patients developing either form 2 or 3 of MERS-CoV infection require intensive medical care in the intensive care unit (ICU). From those, up to 70% typically go to mechanical ventilation [100,101].
It is reported 50% of patients infected by MERS-CoV develop an atypical symptom called Acute Kidney Injury (AKI), which is a sudden kidney failure or damage. From those, up to 70% will pass by renal replacement therapy given the damage caused by MERS-CoV to kidneys [[102], [103], [104], [105]]. MERS-CoV has been detected in upper and lower respiratory secretions at relatively high virus load and fecal samples [106,107].
3.3. SARS-CoV-2
The outbreak of SARS-CoV-2 leading to the world fully spreading the COVID-19 caused more than 2,041,232 million deaths by now [108,109]. After that, many other countries reported increasing COVID-19 cases [110] (Fig. 1). Since WHO declared a pandemic status in March 2020, the global spread rate has accelerated, and confirmed cases are approaching 95,553,377 million infected people [109]. The SARS-CoV-2 belongs to the beta-coronavirus (2B lineage) of the Coronaviridae family, with a positive-stranded RNA genome composed of 29,800 nucleotides in length allowing the production of the same set of proteins seen in other coronaviruses (Fig. 2) [111].
The genome of SARS-CoV-2 is closely related to SARS-CoV-1 (79%) and to a lesser extent of MERS-CoV (50%) (Supplementary Fig. 1B), deadly human coronavirus described in recent years -Coronaviridae Study Group of the International Committee on Taxonomy of Viruses [112]. Although the infection mechanism between these three coronaviruses is similar, the genome sequence reveals some differences [113]. All these epidemics scenarios imposed by coronaviruses have threatened human health, social, and economic context, leading to catastrophic consequences.
3.3.1. Origin and evolution of SARS-CoV-2
SARS-CoV-2 belongs to the Beta-coronavirus B lineage, the same evolutionary branch of SARS-CoV-1 and MERS-CoV, conferring several structural, genetic, and pathogenic characteristics (Supplementary Fig. 1A) [[114], [115], [116]]. Although there is a lack of conclusive evidence, some studies have proposed a plausible explanation for the origin of SARS-CoV-2. The full-length genomic sequence analysis, distinctive phylogenetic distances on the major clade of SARS-CoV-2 provide a clue to the evolutionary relationships among them [116]. However, SARS-CoV-2 shares 79% sequence identity to SARS-CoV-1 and only 50% to MERS-CoV in the genomic sequence. In our genetic distance analysis of the S gene (Supplementary Fig. 1B), SARS-CoV-2 (NC_045512.2) were compared against SARS-CoV-1 (NC_004718.3) and MERS-CoV (NC_038294.1). The evolutionary distance analysis showed considerable differences between SARS-CoV-2 and MERS-CoV, mainly in the N-terminal domain and RBD nucleotide sequence, which are involved in recognizing the cellular receptor (Supplementary Fig. 1B) [117].
Zhou et al. [115] demonstrated SARS-CoV-2 likely evolved from naturally SARS-like coronavirus colonizing bats. Results indicated the closest relationship of SARS-CoV-2 with batCov RatG13 (from Rhinolophus affinis), supporting the hypothesis that Bats are a reservoir for SARS-CoV-2 progenitor. However, although these two coronaviruses are identical (96% of genome identity), RaTG13 S protein diverges in the RBD, suggesting it may not bind to human ACE2 [118]. Pangolins (Manis javanica) was considered the probable reservoir since the pangolin's coronaviruses exhibit high similarity with RBD of S protein from SARS-CoV-2 [119].
Still, neither bat nor pangolin coronaviruses have polybasic cleavage sites, sequence responsible for determining viral infectivity and host range, which raises the question about the possibility of these SARS-CoV-2 progenitors [120]. Mutations, insertions, and deletions can occur near the S1–S2 junction of S protein from SARS-CoV-2, increasing the probability of polybasic cleavage site acquisition, enhancing transmission crossing the mammalian-human line, and improving human-to-human [121] (Fig. 3). Once acquired, adaptation was fixed as a new feature in the SARS-CoV-2 genome and enabled the rapid spread of SARS-CoV-2 to pandemic status.
3.3.2. Pathogenicity
Respiratory air droplets, aerosol, direct contact with contaminated surfaces, and fecal-oral transmission drive human coronaviruses. SARS-CoV-2 reaches the host via the respiratory tract, alveolar epithelial cells, vascular endothelial cells, and alveolar macrophages are the primary targets of the viral entry (Supplementary Fig. 2) [122].
Cell entry of SARS-CoV-1 and SARS-CoV-2 depends on the S protein binding with specific cellular receptors, ACE2 and TMPRSS2, playing a crucial role in the entry of both viruses into the host cells [28]. Briefly, the S protein of SARS-CoV-2 consists of two subunits, the S1 domain, and the S2 domain. SARS-CoV-2 utilizes RBD of the S1 domain to bind to the cellular receptor ACE2, which stimulates the TMPRSS2 to cleavages protein S at the S1 and S2 sites, allowing the cell membrane fusion and viral entry [27] (Fig. 3). Later, the role of S protein from three pandemic viruses in cell entry will be more in-depth discussed.
The clinical symptoms of SARS-CoV-2 infection are similar to SARS-CoV-1 and MERS-CoV infections described, being pneumonia the most described in the studies with abnormal chest CT examinations. However, patients infected with SARS-CoV-2 rarely have significant upper respiratory signs indicating the targeted cell may exist in the lower respiratory tract. Indeed, it has been reported SARS-CoV-2 infection mostly triggers deep airway inflammatory reactions and alveolar damage [123]. For SARS-CoV-2, the most common symptoms are cough, dyspnea, chest pain, myalgia/arthralgia, diarrhea, nausea, vomiting, and common systemic symptoms observed: fever, chills, fatigue [124]. Also, SARS-CoV-2 infects several human tissues, such as the lung, intestinal tract, pharynx, heart, kidney, liver, brain, and blood [125,126]. Nevertheless, further studies are necessary to evaluate the effects of SARS-CoV-2 in extra-pulmonary infection sites.
The immune system may play a critical role in the severity of COVID-19. SARS-CoV-2 infection of pneumocytes induces a “cytokine storm”, which is an activation cascade of auto-amplifying cytokine production [116]. Local inflammatory responses promote the release of cytokines, including transforming growth factor-β1 (TGF-β1), tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), IL-6, IL-2, IL-7, IL-10, granulocyte colony-stimulating factor (G-CSF), monocyte chemotactic protein (MCP), TNF-α and other chemokines to recruit leukocytes for the inflammation region leading to a multiple organ functional failure [116].
A recent study showed a plausible answer for this event: the direct SARS-CoV-2 infection of vascular endothelial cells with concomitant accumulation of inflammatory mononuclear cells in multiple organs in patients with severe COVID-19 [127]. Another plausible point is the involvement of SARS-CoV-2 infection with multiorgan failure due to the ACE2 and TMPRSS2 expression distribution in several human organs [125].
Another aspect is the higher proportion of macrophage and neutrophils observed in the patients with severe symptoms than those with mild symptoms and the chemokine related to these cells [128]. Interestingly, Zuo et al. [129] described the formation of neutrophil extracellular traps (NET) inside the microvessels of severe disease patients that could be a potential factor for severity.
4. Molecular biology of coronaviruses
4.1. Coronaviruses genome organization
Viruses classified as coronaviruses possess a single-stranded, non-segmented, large, and positive-sense RNA. The positive-sense RNA is an advantage acquired over the evolution process that mimics the cellular mRNA making its translation easier. Like cellular mRNAs, coronaviruses RNA possesses both 5′ cap and a tail at 3′ end [130,131] (Fig. 2A). The viral RNA is way larger than cellular mRNA and even compared to other viruses such as picornavirus (four times) and flavivirus (three times). The larger size of the coronaviruses genome varies from 27.3 to 31.3 kb, classified as the bigger RNA molecules are known until now [132].
Coronaviruses genome holds many ORFs (Fig. 2A), another difference compared to cellular mRNA. Precisely, the coronaviruses genome has 10 ORFs. The two largest ORF1a and ORF1b are responsible for producing up to 16 non-structural proteins (nsp) (Fig. 2A). Among those are PL proteinase, 3CL protease, RNA-dependent RNA polymerase (RdRp), and Helicase. The PL proteinase and 3CL protease are involved in polyprotein post-translational processes (Fig. 2A) [67]. The other ORFs are responsible for producing structural and accessory spike glycoprotein (S), an envelope protein (E), membrane attached protein (M), and nucleocapsid protein (N) (Fig. 2B). The genome of coronaviruses produces five canonical proteins. The RdRp at the 5′ end and S, E, M, and N at the 3′ end. This is a common sequence that RNA is translated following the sequence 5′- RdRp-S-E-M-N-3′ (Fig. 2A) [[130], [131], [132]].
Holding a positive-sense RNA is an excellent advantage to any viruses. For coronaviruses, it is not different. If only the RNA entry in a permissive cell the infective process starts. These processes have been shown in many studies over the years [131,[133], [134], [135]]. The most recent example of that is the vaccine produced by companies Pfizer and BioNTech (BNT162b2). BNT162b2 is a lipid nanoparticle–formulated, nucleoside-modified positive sense-RNA encoding membrane-anchored SARS-CoV-2 full-length S protein. Given the positive sense, when RNA is absolved by the cell is promptly translated to viral S protein, which is exposed or externalized by the cells and recognized by the immune system producing antibodies against it [136].
During coronaviruses infection, the genome acts as cellular mRNA producing the larger replicase protein. The RNA from coronaviruses also possesses an important site dedicated to a ribosomal frameshifting event. After that, there is a ribosomal frameshifting site [137] (Fig. 2A). Ribosomal frameshifting in the displacement of ribosomal frames, also known as translation recording, is a biological phenomenon that results in multiple protein production from a single mRNA molecule (Fig. 2A). Although it is common in viruses, it goes beyond them. Organisms from all three kingdoms employ frameshifting to regulate gene expression [[137], [138], [139], [140], [141], [142]]. Next, as usual, the genome is a model for protein synthesis, RNA replication, and assembly, producing new virus particles [143].
4.2. Virion particles
Coronavirus particles are spherical, pleomorphic (Fig. 2B) with diameters varying from 80 to 120 nm [144,145]. Attached to it membranes are found two types of the spike. One larger with projection 17–20 nm long produced S protein (Fig. 2B), and a smaller one 5–10 nm long today known as the hemagglutinin-esterase (HE) found in a subset of coronaviruses [144,145]. Recently, the development of advanced microscopy techniques has clarified coronavirus morphology.
Ng and collaborators [146] catch three-dimensional (3D) images from SARS-CoV moving out from the Vero infected cell. By employing scanning electron (SEM) and atomic force (AFM) microscopies, it was possible to notice the small cauliflowers-like structures assumed by viral particles. To see the outside of the virus is not that hard. The problem is to evaluate the inside of a viral particle. To go that far, virion particles have been treated to nonionic detergents, allowing them to look inside the coronavirus particles. It was revealed that coronaviruses possess helical symmetric nucleocapsids, which is not common to positive-sense RNA viruses but for negative-sense RNA viruses. This is a great point of discordance among virologists. However, a few published studies are reporting that, but only we know is that more studies are required to obtain a clear picture of coronavirus virion particles.
5. The coronaviruses proteins
5.1. Membrane protein (M)
The M protein (Fig. 2B) is one of the three main structural proteins in coronaviruses. It is responsible for the viral particle conformation after assembling [[147], [148], [149], [150]]. The M protein holds a small N-terminal domain extended to the outside of the viral particle, which is important to interact with the endoplasmic reticulum during infection [151]. Right after the N-terminal domain, there are three large, highly hydrophobic transmembrane domains followed by an even larger C-terminus domain, which stands inside the viral particle.
The M protein has regular conservation into the coronavirus family. The same is not applied to the coronaviruses groups. A consensus in all groups is that M protein has multiple-glycosylated stages. The preglycolysated stage of M proteins ranges in size from 25 to 30 kDa. However, multiple glycosylations could lead to a high molecular Mass of M protein [152]. Neuman et al., [147] reported the ability of M protein to oligomerizes to form larger structures. In the same study, the authors revealed two stages: the larger and compact. In the larger stage, M protein forms a structure 8 nm long. In contrast, the compact form has structures 6 nm long. The oligomerization stages of M protein are involved in many virion particle assemblies and even genome protection [147,[153], [154], [155]]. The interaction between transmembrane domains drives the interaction between M proteins; this interaction is important to exclude any leftover host membrane during the release of viral particles [147,156].
5.2. Envelope protein (E)
The E (Fig. 2B) is the smallest, membrane-integral, and less known structural protein from coronaviruses. One quite intriguing point is that E protein accumulates the most in the infected cells during viral replication. Still, only a very small portion is inserted in the new viral particle. The function of the excess of E protein in replication is obscure [157,158]. Most of E protein is localized outside the viral particle. This is because it is involved in the viral attachment to the ER and Golgi apparatus controlling virus intracellular traffic [[158], [159], [160]]. Although not known how the E protein is involved in the viral titers, viral particle production, and maturation [159,[161], [162], [163], [164], [165], [166], [167], [168]].
5.3. Nucleocapsid protein (N)
The N (Fig. 2B) protein holds a molecular weight varying from 43 to 50 kDa, larger than M and E proteins but far smaller than S protein. It has a quite essential function to coronaviruses. Interacting with the RNA genome forms a helical structure like a necklace with beads providing stability to the RNA genome [169]. N protein sequence is divided into three domains. The highest part of the molecule has great amounts of Lys and Arg residues leading to a positive charge, which is involved in the interaction with the negative charge of viral RNA. This portion comprehends domains 1 and 2. For example, it was identified that the RNA bind ability of MHV N is attributed to domain 2 [[169], [170], [171]].
An unusual feature of N proteins in the phosphorylation. N protein from some but not all coronaviruses are phosphorylated. Some phosphorylation happens in a Ser and others in the N residues [172,173]. The biological function of phosphorylation in the N until today is obscure. However, the hypothesis is that the phosphorylation helps N protein to recognize only viral RNA. Chen et al. [172], employing Mass spectroscopic analysis and surface plasmon resonance, brought some information about the biological importance of N phosphorylation. The nonphosphorylated and phosphorylated N protein has the same ability to bind to viral RNA. However, the phosphorylated N protein binds only to the viral RNA. In contrast, the nonphosphorylated binds to both viral and non-viral RNA. These experiments by Chen et al. [172] revealed one biological effect of phosphorylation of N protein, which is the recognition exclusively of viral RNA.
5.4. Spike protein (S)
Now, after a brief explanation about coronaviruses and their proteins. It will be deeply discussed about the protein that is the focus of the review and plays essential roles during coronaviruses infection, Spike glycoprotein (S). The S protein is the third protein component and most abundant in the viral envelope (Fig. 2) [174]. The S protein is the larger protein from coronaviruses with about 180–200 kDa [174]. It has a larger N-terminal region (90% of protein) exposed outside the viral envelope and a shorter C-terminal region inside the viral envelope. Between N- and C-terminal regions, there is a transmembrane section liking both N- and C- regions (Fig. 3A) [174].
The S protein possesses high-level glycosylation on its structure. Before post-translational processing, the monomeric structure of S protein that inserts carbohydrates on its structure is about 128–160 kDa. After the glycosylation process, the size of S protein ranges between 180 and 200, which indicates a high amount of carbohydrates are inserted in its structure, most of them N-linked [175]. It is proposed that the biological function of glycosylation in the S proteins is to evade the host immune system during infection [176,177]. For example, a study from Delmas and Laude [177] with the S protein from porcine transmissible gastroenteritis virus revealed the glycosylation process coincides with the translation. The authors also showed that the glycosylation precedes the S protein trimerization. The glycosylation of TGEV S is essential to correct the folding of monomers [177].
S protein is still a huge protein even in the monomeric stage with 1273 amino acid residues. The first 13 amino acids form the signal peptide in the N-terminal site targeting the protein from the endoplasmic reticulum to the viral envelope. The S1 subunit goes from 14 to 685 amino acid residues. The S2 subunit starts at the amino acid 686 until the 1273 residue. Both regions are involved in cell recognition and entry (Fig. 3A and Supplementary 3) [178,179]. Like other proteins, the S protein is synthesized as an inactive precursor present in the viral envelope. During cell infection, the S protein is cleaved by a cellular protease releasing the S1 and S2 subunits activating it and allowing the membrane fusion and virus entry (Fig. 3A) [28,180,181]. This will be discussed later in this review.
5.4.1. The S1 subunit from S protein
The S1 subunit is smaller than the S2 subunit but has a quite relevant coronaviruses function (Fig. 3A and Supplementary 3). The S1 subunit is responsible for recognizing the cell receptor to allow virus entry into the cell cytoplasm. The S1 subunit hosts the RBD responsible for binding to ACE2 in the portion where the aminopeptidase cleavage point is present [[181], [182], [183]]. The S1 region also contains the N-terminal (NTD) and C-terminal (CTD) domains, both involved in the RBD recognition (Fig. 3A and Supplementary 3). For comparison purposes, the SARS-CoV-2 S1 CTD has more amino acid residues (21 aa) directly involved with the ACE2 interaction that SARS-CoV S1 CDT does (17 aa). Additionally, the mutational analysis revealed amino acid substitution from I472 in SARS-CoV S1 CTD for F486 SARS-CoV-2 S1 CTD strength interaction with the ACE2 Y83, the establishment of aromatic-aromatic interaction (15, 16). Yet, the substitution of E484 in SARS-CoV-2 S1 CDT instead of a P470 residue in SARS-CoV S1 CTD increases the ionic interaction with ACE2 leading to higher affinity to the receptor (Fig. S3) [182,183].
The RBD hosted by the S1 domain is critical for recognizing the ACE2 receptor by S protein. There are nine residues fully conserved in all coronaviruses involved in the ACE2-RBD interaction. Given the importance of RBD for the coronavirus, it becomes a great target for the action of neutralizing antibodies [27,28,180,182,183]. Our research group has recently performed a molecular docking with synthetic peptides against S protein from SARS-CoV-2 [184]. Out of eight, two peptides showed up as promising the most to bind toward S protein. Indeed, the peptides do not interact with the RBD domain. However, the interaction with S protein led to conformational changes in the S protein structures. The docking simulation led to a wrong interaction with the ACE2 receptor, hence inhibiting virus entry in the cell [184]. In another study, Souza et al. [185] designed four peptides from the amino acid sequence of the ACE2 receptor. Those peptides specifically targeted the RBD domain from S protein. After interaction with peptides, RBD presented changes in the conformational structure impairing the correct interaction with ACE2 protein, suggesting that peptides could prevent cell infection by SARS-CoV-2.
5.4.2. The S2 subunit from S protein
The S2 subunit from the S protein hosts many important domains for S protein function. The first domain is the fusion peptide (FP) from 788 to 806 amino acid residues (Fig. 3A and Supplementary 3). FP is a short fragment highly conserved in the coronavirus family. Its composition is essentially made by apolar residues such as Gly and Ala, all-important during the membrane fusion activity of the S2 subunit during viral infection (Fig. 3A and Supplementary 3) [186] [13,23].
Right after the FP domain, two sequences called heptapeptide repeated sequence 1 (HR1) (912–984 residues) and heptapeptide repeated sequence 2 (HR2) (1163–1213) with, respectively, 72 and 50 (Fig. 3A and Supplementary 3) [187]. Both HR1 and HR2 are located at the N-terminus portion of the transmembrane domain (TM) and follow the same structure HPPHCPC where H designates a hydrophobic residue, P regards a polar or hydrophilic, and C is any other charged residue. The HR1 and HR2 are essential to the viral fusion and entry function of the subunit in the cell [187].
The HR1 domains assemble to form a homotrimeric structure with highly conserved hydrophobic surfaces. These hydrophobic surfaces are exposed to the outside of the 3D structure of the S2 subunit to interact with the HR2 domain. In turn, HR2 forms a rigid helix followed by a highly flexible loop in the face that interacts with the HR1 domain. The interaction between HR1 and HR2 domains is strong and supported by hydrophobic and aromatic interactions forming a bigger domain fusion core region [188].
The HR1 and HR2 are essential to infection establishment by hCoVs and are thus highly conserved [188]. Given that, these domains are targeted continuously by potential antiviral molecules. That happens because another critical target, the RBD from the S1 domain of S protein, is highly variable, so it is hard to develop a drug toward RBD. But the high conservation of HR domains makes them a great target [[186], [187], [188], [189]]. For instance, Xia et al. [189] have developed an HR2-derived peptide that targets the HR1 region and inhibits cell infection by human coronaviruses.
6. Biological roles of the S protein
The S protein is the most important factor involved in viral infection. Because of that, it stands exposed on the viral envelope surface. It is a trimetric protein essential to receptor recognition, cell attachment, membrane fusion and virus entry [28,38,54,80,83,85,126,179,182,184,191]. Given S protein roles, it is present in all kinds of hCoVs and other viruses such HIV and Ebola [191] but with other names.
The S protein possesses two conformational states (Fig. 3B). The conformational dynamics of S protein is due to the flexibility of NTD and RBD. The first is the closed state where the RBD domain cannot recognize the ACE2 receptor (Fig. 3B). The closed state is also called perfusion state because it precedes membrane fusion [192]. The closed state (prefusion) of beta coronaviruses was determined by cryo-electron microscopy [193]. In general, the closed state of S protein from coronaviruses is similar to the, considering higher complexity, hemagglutinin from influenza virus. In the closed state conformation, the RBD domain is trapped into a pocket formed by the NTD region and RBD itself, inhibiting the interaction with the ACE2 receptor [85,193].
Additionally, in the closed state, the S1 heads stand on top of S2 subunits to avoid the S2 conformational transitions. In the S2 domain, the HRs domain assumes the helices form to stabilize the S2 subunit. In the coronavirus fusion HR-1 domain, the hydrophobic residues support the formation of a small loop and helix, which are buried inside the prefusion structure. The proteolysis is essential for the conformational transition of S2, one at the S1/S2 border and the other at the N-terminal region [194,195]. The S protein moves to the second stage called an open state (Fig. 3B). In the open state, the RBD is now exposed outside the S protein structure and can recognize and bind to the cellular receptor (Fig. 3B) [27,28,67]. In this stage, the RBD dynamics is modulated by proteolysis allowing the interaction with the ACE2 receptor. After interaction by RBD-ACE2, the virus will start the process to enter within the cell, and the S protein starts to change to another stage called post-fusion, leading to the viral and cellular membrane fusion [27,28,67,189,[193], [194], [195]]. Recently, Wang et al. [196] reported the S protein could have an alternative route to enter cells. Rather than interact with RBD, S protein from hCoVs could interact with a CD147, which is a transmembrane glycoprotein from the immunoglobulin superfamily. It is known for the CD147 involvement in tumor development, bacterial, virus, and Plasmodium invasion and infection [[196], [197], [198]].
Wang et al. [196] revealed Vero E6 and BEAS-2B cell lines either lacking or blocked to CD147 or blocking presented no signal of SARS-CoV-2 amplification. Additionally, the expression of CD147 in SARS-CoV-2 non-permissive human cells allowed virus entry and infection. The authors also found high viral loads in the mice lungs expressing human CD147, but not in wild type mice. Indeed, the results presented by Wang and co-authors [196] are quite interesting.
In the same way, since November 2020, a new cellular receptor came up as hosts in facilitating SARS-CoV-2 entry. Those receptors either enhance the ACE2-mediated entry of SARS-CoV-2 or act as alternative receptors for SARS-CoV-2 entry. SARS-CoV-2 could employ these alternative receptors to enter the cell with a low expression of ACE2. For instance, the tyrosine-protein kinase UFO (AXL) acts as a receptor by interacting with the NDT domain of S protein, demonstrated by in vitro cell culture model and COVID-19 patients samples [201]. Another in vitro model study showed the high-density lipoprotein (HDL) scavenger receptor B type 1 (SR-B1) as a facilitator of ACE2-dependent entry of SARS-CoV-2 through the interaction of the S1 subunit. Blocking the S protein interaction with a specific monoclonal antibody against cholesterol/HDL inhibits the HDL-enhanced SARS-CoV-2 infection [202].
Cantuti-Castelvetri et al. [203] and Daly et al. [204] revealed that neuropilin-1 (NRP1), which is known to bind furin-cleaved substrates, potentiates SARS-CoV-2 infectivity. The NRP1 protein is highly expressed in the respiratory and olfactory epithelium, with the highest expression in endothelial and epithelial cells. Those cells present a low expression of ACE2 receptors, which is why SARS-CoV-2 employs this protein to enter the cell [203,204]. Amraei et al. [205] reported that CD209L and CD209 are members of the C-type lectin superfamily and could act as mediators of SARS-CoV-2 entry cell and increase viral pathogenesis. The CD209L is highly expressed in human type II alveolar cells, lungs, and liver, whereas CD209 protein is expressed in alveolar macrophages. Both CD209L and CD209 interact with the RBD of S protein from SARS-CoV-2 to enter cells. The mechanism of these proteins is quite similar to ACE2-mediated cell entry by SARS-CoV-2.
Recently, in an elegant experiment, Tang et al. [206] revealed a new candidate molecule used by SARS-CoV-2 to enter cells. ACE2-knockout mice are not protected from SARS-CoV-2 infection. The authors showed that the transferrin receptor (TfR) is the target by S protein to allow SARS-CoV-2 entry on cells. Even though new finding supports alternative molecule employment to invade cells by SARS-CoV-2, little is known about the role of these receptors for SARS-CoV-2 entry on cells. Based on that, the discussion below will be mainly focused on the ACE2-S protein interaction, which there more information allowing a deeper discussion has been supported by the literature.
Most of the CoVs enter cells by interacting with the RBD domain from S protein with the ACE2 receptor [199]. The ACE2 receptor is a widely expressed protein in the cell membrane of many tissues such as lungs, heart, kidney, renal, cardiovascular, and intestine [200]. Discovered in 2000, ACE2 holds 61% of similarity with ACE. Structural analysis of ACE2 protein revealed an N- and C-terminal followed by a unique transmembrane ɑ-helix domain followed by an intracellular portion [113]. The difference between ACE and ACE2 is the most related to their activity. At the time, ACE catalyzed the conversion of angiotensin I in angiotensin II. ACE2 acts in angiotensin II conversion in two forms: angiotensin 1–1 and angiotensin 1–9 [201]. Until the pandemic situation imposed by the coronaviruses, ACE2 moves from the shadow to light as an important protein to viral infection. Now, it is quite clear that SARS-CoV-2 and other coronaviruses employ the ACE2 to come into a cell and start an infection.
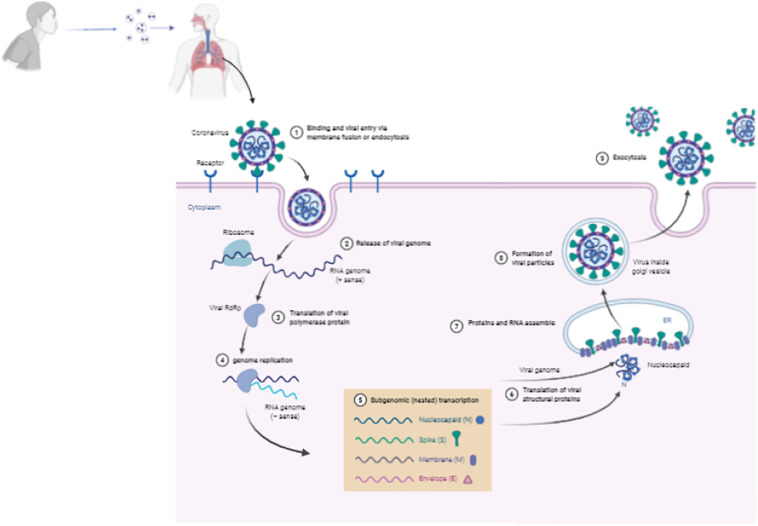
The infectious process starts when a coronavirus-infected person sneezes, releasing air droplets containing virus particles into the environment (Fig. 4 ). Without a social distancing, a healthy person intake, by natural breathing, the contaminated air allowing the air droplets infected with the virus to reach the lungs (Fig. S2). At that point, the S protein becomes the principal actor in the coronavirus infection by driving viral recognition and entry on cells. The S1 subunit from the S protein starts the process by promoting RBD interaction with ACE2 (Fig. 4 - 1). After the attachment of the virus supported by S protein and low pH conditions act as targets to the viral fusion of membranes [178,179,182].
Fig. 4.
Schematic model of coronavirus entry in cell. Coronaviruses bind to the host cell surface and release their RNA genomes into the cell through endocytosis. The positive-sense RNA genome is translated to produce the RNA-dependent RNA polymerase (RdRp) complex. Then the RdRp complex produces negative-sense RNA from the RNA genome, which provides the template for synthesis of positive-sense mRNAs. Then these subgenomic mRNAs are translated into structural and accessory proteins for viral particle assembly in endoplasmic reticulum. Finally, The enveloped virion is then exported from the cell by exocytosis. Created with BioRender.com.
After the attachment, the S protein, driven by the S2 subunit, promotes the viral fusion, which is the fusion of the viral membrane with the host cell membrane. The trigger to initiate the membrane fusion is the proteolytic action suffered by S protein after the attachment to the receptor. As a result of the cleavage, the S protein is separated into two parts, S1 and S2, by cellular proteases. Even after the cleavage, the subunit S1 and S2 remain non covalently bonded to ensure the membrane fusion follows through [202,203].
Hasan et al. [203] and Millet et al. [202] revealed that in the SARS-like coronaviruses such as SARS-CoV and SARS-CoV-2, the S protein has an uncleaved state. In contrast, other coronaviruses possess S protein mainly in the cleaved state. Additionally, the mutational analysis revealed that SARS-CoV-2 had accumulated mutations increasing the number of cleavage sites on S protein, leading to higher cleavage rates and cell entry and infectivity [202,203]. Many studies over the years [28,180] revealed host proteases involved the most in S protein cleavage are TMPRSS2 and trypsin, even for SARS-CoV and SARS-CoV-2. The S protein from SARS-CoV and SARS-CoV-2 are essentially the same. However, the high number of cleavage sites in the SARS-CoV-2 S protein might be involved in higher contagious levels of SARS-CoV-2 compared to SARS-CoV.
Back to viral fusion, the domains FP and both HR on S2 is critical to viral fusion. The cleavage of S protein exposes the FP domain, which is, in turn, inserted in the host membrane triggering the viral fusion [114,178,191]. Once anchored in the host membrane by the FP domain, the distances between viral and host membrane are dramatically reduced, and HR1 stands close to the cell membrane, and HR2 is yet anchored in the viral membrane. Now starts the last and crucial process in membrane fusion. The HR2 domain turns up toward the HR1. Both domains form a helix structure to produce a fusion core, and the viral membrane is pushed over toward the cell membrane tightly, and both membranes fuse. The virus reaches the cytoplasm. Now it is just a matter of making the cell work in virus replication. Once inside the cytoplasm, the virus hijacks cellular proteins and supplies to replicate the genome and produce thousands of new viral particles [204,205].
Inside the cell, the positive-sense RNA from coronaviruses, which works as mRNA, is released, and translated by the cellular ribosomes (Fig. 4 - 2). The first protein produced is the huge replicase polyprotein complex (Fig. 4 - 3). The positive-sense RNA is then used as a model by the replicase to produce the negative-sense RNA, the subgenomic RNAs, and thus new positive-sense RNA molecules (Fig. 4 - 4). The subgenomic RNAs were used to produce S, E, M, N proteins (Fig. 4 - 5 and 6). The proteins S, E, and M move to the ER and attach to its membrane. At the same time, the N protein interacts with the RNA molecules to form the nucleocapsid particles (Fig. 4 - 7). Now, induced by structural membrane-bounded protein, the membrane of ER suffers conformation changes to assume a spherical format with the nucleocapsid particle inside, which is the new virion particle. The new viral particle moves now out to infect new cells (Fig. 4 8–9). This is the common way of CoVs replication. S protein is not called here as a principal actor without a motive. In some cases, during CoVs, the infection can lead to the formation of syncytia [[206], [207], [208]].
Syncytia are giant infected cells induced by some viruses, like coronaviruses. All pandemic coronaviruses, SARS-CoV-1, MERS-CoV and SARS-CoV-2 [[206], [207], [208]] have the ability to induce syncytia formation. Somehow, in coronavirus infection, the host cell showed the S protein on the membrane, which is a quite important function of S protein. The S protein interacts with the ACE2 with neighboring healthy cells and thus induces membrane fusion of many cells forming giant known syncytia. By inducing the syncytia formation, the coronaviruses can continuously infect cells without moving out cells. This is an incredible and essential ability acquired by S protein over evolution. The syncytia formation induced by S protein allows the virus to keep spreading without extracellular virus particles working as an evasion mechanism of the immune system.
7. S protein in the pandemic coronaviruses
As discussed above, in the last two decades, the human king faced three outbreaks caused by hCoVs. These coronaviruses have threatened and taken many lives worldwide with different spreading rates, fatality cases, and regions affected (Fig. 1 and Table 1). The pandemic hCoVs had present different spread and human-to-human transmissible rates. That factor could be attributed to the mutation in the S protein from all of them. Over the years, hCoVs have accumulated many mutations, some negative, some positive, which culminate in the current pandemic scenario. This section will discuss the characteristics and differences of S protein from the pandemic hCoVs. Most of the comparisons are made between SARS-CoV and SARS-CoV-2 when pertinent MERS-CoV will be included.
The mutations that lead to the change in amino acid in the S protein could be important information to track the natural sources of CoVs, allowing intra-species or inter-species jumping of the virus. For example, genomic sequence experiments revealed that SARS-CoV-2 is closer to bat CoVs than other hCoVs, which indicates its origin [37,51,62,69,115,117,121,190,209]. The RatG13, CoV from bats, has a similarity of 93.1% in the gene and 98% in the protein sequence of S protein with SARS-CoV-2. In contrast, SARS-CoV-2 has only 80% similarity in the S protein with other hCoVs [37,51,62,69,115,117,121,190,209].
In contrast to other CoVs, SARS-CoV-2 has accumulated many mutations in S protein conserved regions (Fig. S3). Because of that, even with some similarities, SARS-CoV-2 is much sufficiently different from SARS-CoV and SARS-like coronaviruses [40,47,51,111]. In comparison to pandemic hCoVs, SARS-CoV-2 is, indeed, closer to SARS-CoV than MERS-CoV (Fig. S1A) [45,93,98,178]. Even though evolutionary studies put SARS-CoV-2 and SARS-CoV in the same group, they still hold remarkable variations between them [111]. Wu et al. [111] revealed high similarities between SARS-CoV-2 and SARS-CoV at the polyprotein level, but many significant alterations exist. For instance, the most remarkable is the absence of protein 8a in SARS-CoV-2 and its presence in the SARS-CoV, which indicates a mutation that deletes or produces a truncated inactive 8a protein in SARS-CoV-2. For sure, the absence of 8a did not affect SARS-CoV-2 fitness [111].
To have a better picture of the S protein changes of the pandemic hCoVs, we performed an alignment comparing the entire sequence of S protein from MERS-CoV (accession number K9N5Q8), SARS-CoV-1 (accession number P59595), and SARS-CoV-2 (accession number P0DTC2) deposited in the UniProt database (Supplementary Fig. 3). By analyzing the results, the first point noticed was the length of S protein in all viruses. The MERS-CoV S protein has a higher number of amino acid residues, 1345. In contrast, S protein from SARS-CoV and SARS-CoV-2 has, respectively, 1243 and 1261 amino acid residues (Supplementary Fig. 3). That might be a result of many deletions in S protein from both SARS-CoV and SARS-CoV-2.
The S protein alignment from the pandemic hCoVs revealed that compared to the MERS-CoV S protein, the S protein from SARS-CoV-1 and SARS-CoV-2 has 14 points of deletions. Probably, because of those deletions, the S protein from SARS-CoV-1 and SARS-CoV-2 are smaller than the MERS-CoV S protein (Supplementary Fig. 3). Of these, 12 points of deletions are present in the S1 subunit and 2 on the S2 subunit. Most of the point mutations are situated in regions that have no important domains for the infection process. But somehow alter the 3D of S protein, making it more compact in both SARS-CoV and SARS-CoV-2 than MERS-CoV.
Another great difference between S protein from SARS-CoV and SARS-CoV-2 is the pattern of glycosylation [[210], [211], [212]]. The presence of an additional glycosylation spot in Asn370 in SARS-CoV-2 compared to S protein from SARS-CoV provides an additional glycosylation state in S protein from SARS-CoV-2. This spot belongs to a domain involved in the membrane attachment and thereby enhances the interaction with the receptor and membrane fusion [211]. Point mutations in SARS-CoV-2 S protein are said to increase virulence through the instability of the viral machinery and altered viral-to-cell-membrane fusion (Supplementary Fig. 3).
It is already known that the amino acid composition change does not reflect protein structure changes and thereby function. To check that, we provide a root mean square deviation (RMSD) calculation of the atomic position on S protein from the pandemic hCoVs (Fig. 5 ). The RMSD provides a value: If the value is 0, the atoms from both structures are in the same position, with no alteration in the 3D structure. However, if the value is anyone higher than zero, it indicates the atoms are in different positions, and this value is high, the structures are very different.
Fig. 5.
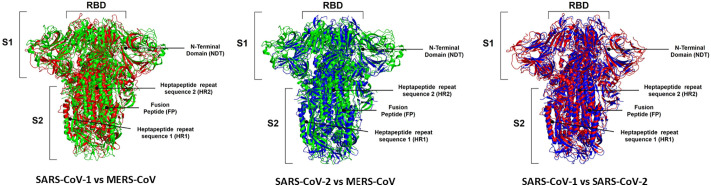
Three-dimensional structure comparison of Spike protein among SARS-COV-1, MERS-CoV and SARS-COV2 through protein alignment. The spike protein of SARS-CoV-1 is represented in red, MERS-CoV in green and SARS-CoV-2 is blue. The 3D structures from S proteins MERS-CoV (PDB ID: 5X5C), SARS-CoV-1 (PDB ID: 5X58), and SARS-CoV-2 (PDB ID: 6Z97) deposited in the UniProt database deposited in the Protein Data Bank (PDB, https://www.rcsb.org/). RBD: Receptor-binding domain; NDT: N-terminal domain; HR1 and HR2: heptapeptide repeat sequence 1 and 2; FP: fusion peptide of subunit 1 and 2 are identified in the images. The structural alignments were performed in the Pymol program with the educational license.
To perform the structural alignments was used the 3D structures from S proteins MERS-CoV (PDB ID: 5X5C), SARS-CoV-1 (PDB ID: 5X58), and SARS-CoV-2 (PDB ID: 6Z97) deposited in the UniProt database deposited in the Protein Data Bank (PDB, https://www.rcsb.org/). By performing a structural alignment, it is possible to see structural changes. Fig. 5 shows the structural alignment between the S proteins from MERS-CoV and SARS-CoV. It is possible to analyze in many positions the differences in both proteins. The RMSD value for this alignment was 12.93 A, indicating both atomic positions are quite different. The comparison between S protein from MERS-CoV and SARS-CoV-2 is more interesting (Fig. 5). In some regions such as NDT, the arm of S1 and HR is possible to evaluate that the S protein structure from MERS-CoV is more prominent than S protein than SARS-CoV-2. It seems the S protein from SARS-CoV-2 is more compact with a lower atomic distance than MERS-CoV (Fig. 5). The RMSD value for the structural alignment of S protein from MERS-CoV and SARS-CoV-2 is 15.90 A, which indicates both structures are quite different. Despite the differences in S protein it is important to remember both viruses employed different receptor to come inside the cell. Together these differences are responsible for complete behaviors regarding the spread, infectivity, and severity of the disease.
The RMSD analysis of S protein from SARS-CoV and SARS-CoV-2 (Fig. 5) revealed a lower value, 9.21 A, than that obtained in the comparisons of SARS-CoV x MERS-CoV and SARS-CoV-2 x MERS-CoV. It is important to notice that even the S protein from SARS-CoV and SARS-CoV-2 are similar, they share many differences. By looking at amino acid sequence alignment (Supplementary Fig. 3) it is possible to notice many changes in the amino acid residues at the same position. These changes could be responsible for the altered structures revealed by RMSD analysis.
It is important to notice that some point mutations occur on the RBD site (Supplementary Figs. 3 and 4). To have a better overview it was performed an alignment comparing the sequence of RBD extracted from the sequence of S protein from MERS-CoV (accession number K9N5Q8), SARS-CoV-1 (accession number P59595), and SARS-CoV-2 (accession number P0DTC2) deposited in the UniProt database (Supplementary Fig. 4). For example, the sequences 536EDCDYYRKQLS546, 575VQYC578, and 587KLEFAN592 are present in the S protein from MERS-CoV absent in that one from SARS-CoV and SARS-CoV-2 (Supplementary Fig. 4). Deletion points were not an exclusivity from SARS-CoV and SARS-CoV-2. The S protein from MERS-CoV presents 4 points of deletion than SARS-CoV and SARS-CoV-2 (Supplementary Fig. 3).
The RBD portion has nine cysteine residues able to form four disulfide bonds. Comparing the location of these disulfide bonds in the SARS-CoV, MERS-CoV, and SARS-CoV-2, it was possible to see if the positions of disulfide bonds are similar SARS-CoV-2 and SARS-CoV, but in a different position in comparison both with MERS-CoV (Fig. 5 and Supplementary Fig. 3) [183]. This result suggests a closed mode of action between RBD from SARS-CoV and SARS-CoV-2 but not too close to the action of RBD from MERS-CoV.
Regarding S protein, most of the amino acid changes were in RBD, affecting the conformational flexibility of the protein or binding interactions with ACE2 [213]. The mutations accumulated by SARS-CoV-2 lead to five amino acid residues that are different than those in SARS-CoV. In SARS-CoV, the residues are Tyr455, Leu486, Asn494, Asp495, Tre501, and Tyr506. In contrast, in SARS-CoV-2, the residues are Leu455, Phe486, Glu494, Ser495, Asn501, and Tyr506 [214] (Supplementary Fig. 4). These differences in the SARS-CoV-2 RBD domain allow it to bind to ACE2 with an affinity 20 times higher than SARS-CoV [214].
A key mutation in the RBD from SARS-CoV-2 compared to SARS-CoV is the presence of a Lys417. The core region of RBD from SARS-CoV-2 has a unique residue of Lys417, forming a salt-bridge interaction with a residue of Asp30. This interaction strengthens the energy bind of SARS-CoV-2 RBD with ACE2. In contrast, this position of RBD from SARS-CoV holds a Val residue that did not allow such type of interaction (Supplementary Fig. 4) [183]. Indeed, the presence of Val residue interrupts the interaction, thus weakens the interaction with ACE2. Besides, the presence of Lys417 provides a positively charged patch providing an electrostatic surface, which is not present in the RDB from SARS-CoV [183]. Besides strengthening the SARS-CoV-RBD::ACE2 interaction, the Lys417 is also important to S protein immunogenicity (Supplementary Fig. 4). The substitution of a Val to Lys in the RBD from SARS-CoV-2 hinders the interaction of anti-SARS-CoV antibodies and thus the neutralization of SARS-CoV-2 by anti-SARS-CoV antibodies (Supplementary Fig. 4) [183].
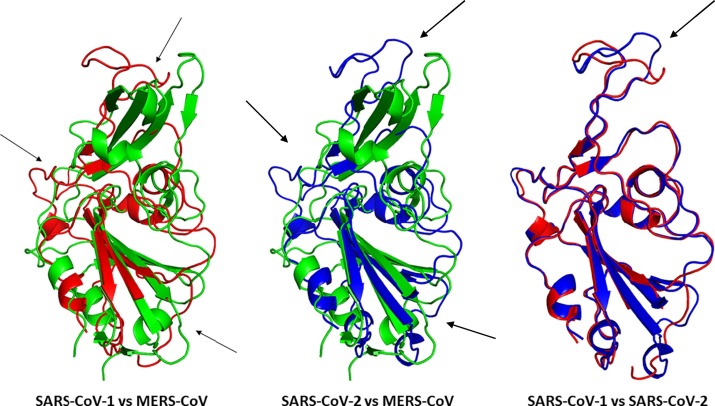
The RMSD analysis revealed some important information about the structure of all S proteins from pandemic hCoVs. However, the S protein is a huge protein with a complex structure (Fig. 5). To obtain a better view of the alteration in RBD, it was performed an RMSD analysis of only the RBD portion, comparing in all pandemic hCoVs. The RMSD allowed a more in-depth evaluation of differences (Fig. 6 ). To perform the structural alignments was used the 3D structures of RBD from MERS-CoV (PDB ID: 6L8Q), SARS-CoV-1 (PDB ID: 2AJF), and SARS-CoV-2 (PDB ID: 6VW1) deposited in the UniProt database deposited in the Protein Data Bank (PDB, https://www.rcsb.org/). The comparison between RBD from MERS-CoV and SARS-CoV revealed at least three positions with a high variation in the structure (Fig. 6 arrows). The RMSD value was 10.34 A. Corroborating the results from S protein RMSD analysis of MERS-CoV and SARS-CoV-2 (Fig. 6), the RBD analysis domain presented a high RMSD value of 11.6 A, indicating the structures were quite different. The arrows highlighted the most prominent differences (Fig. 6).
Fig. 6.
Three-dimensional structure comparison of Receptor-Binding Domain (RBD) of Spike protein among SARS-COV-1, MERS-CoV and SARS-COV2 through protein alignment. The RBD of SARS-CoV-1 is represented in red, MERS-CoV in green and SARS-CoV-2 is blue. Structural differences are indicated by arrows. To perform the structural alignments was used the 3D structures of RBD from MERS-CoV (PDB ID: 6L8Q), SARS-CoV-1 (PDB ID: 2AJF), and SARS-CoV-2 (PDB ID: 6VW1) deposited in the UniProt database deposited in the Protein Data Bank (PDB, https://www.rcsb.org/). The structural alignments were performed in the Pymol program with the educational license.
Regarding the RMSD analysis of RBD follows the same pattern of S protein for SARS-CoV and SARS-CoV-2 (Fig. 6). Although there are many differences in the amino acid sequence of RBD showed by the alignment (Fig. S4). The conformational structure of RBD from SARS-CoV and SARS-CoV-2 is quite similar as represented by the RMSD analysis with a value of 2.09, which is very low compared to other RMSD analysis (Fig. 6). The similarity between RBD sequences from SARS-CoV and SARS-CoV-2 is around 74% allowing both viruses to use ACE2 receptors to a different extent in the cell [118]. Indeed, RBD from SARS-CoV-2 can form a higher number of interactions with ACE2 than RBD from SARS-CoV, supporting the high affinity of SARS-CoV-2 RBD to ACE2 than SARS-CoV. This is possible because RBD from SARS-CoV-2 performs more atomic interaction with the ACE2 compared to SARS-CoV. Experiments have shown the RBD from SARS-CoV-2 binds to ACE2 in an nM scale concentration, with dissociation at 4.7 nM. In contrast, the RBD from SARS-CoV has a constant of dissociation about 31 nM [28,178,183].
All these differences in the atomic arrangement led to suitable differences in S protein are directly involved in the interactions with the ACE2 receptor and thus binding affinities. Based on that, it is feasible to suggest that those alterations are responsible for the higher affinity of SARS-CoV-2 RBD than SARS-CoV RBD by the receptor, and thereby higher transmissibility [28,178,183]. Experimental analysis using animals is an important source of information if the mutations on RBD affect COVID-19 symptomatology and may provide information about the origin of the pandemic situation [215]. Based on RBD sequences' similarities, there have been discussions of convergent evolution between SARS-CoV and SARS-CoV-2 RBD structures enhancing the affinity of SARS-CoV-2 by ACE2 [183,215].
Albeit everybody looks for a mutation in the RBD given its interaction with ACE2 receptor, other regions. For example, it was reported that many mutations changed amino acid sequences in the HR1 domain of SARS-CoV-2 compared to SARS-CoV, which might be associated with improved and more robust interaction with the HR2. Thereby enhancing the membrane fusion ability of S protein, which increased SARS-CoV-2 infectivity (Fig. S3) [181,189,193,195]. Comparing SARS-CoV and SARS-CoV-2, the S2 domain is well preserved in both N- and C-terminal domains. All regions involved in viral particle fusion such as FP, HR1, HR2, TM, cytoplasmic portion have similarities, respectively, of 93%, 88%, 100%, 93%, and 97% (Fig. S4) [216].
It was already described how mutations favoring SARS-CoV-2 S protein cleavage improve viral loads and the higher efficiency of spread compared to other hCoVs [27,181,216]. These mutations increase the number of proteases that could cleavage the extracellular portion of S protein from SARS-CoV-2, increasing its infectivity and virulence [118,181,213]. Mutational studies showed that compared to other coronaviruses S protein, one from SARS-CoV-2 has a higher number of hotspots for glycosylation. It is quite possible that SARS-CoV-2 is using these new sites of glycosylation to evade immune system surveillance, making it S protein less antigenic than those from other coronaviruses do and thereby contributing to its pandemic spreading [118,181,207,213,217,218].
8. Mutations in S protein from SARS-CoV-2
Based on all discussion made above, it is quite clear the high mutational rates in hCoVs. Most of the discussion focused on the mutations in the pandemic hCoVs SARS-CoV, MERS-CoV, and SARS-CoV-2. In this topic, the focus will be the mutations that happened in the S protein from SARS-CoV-2 since the beginning of the outbreak.
Mutations are a process intrinsic to all RNA viruses. There are some results from mutation: (1) mutations are a natural process resulting from the errors made by the RNA polymerase during RNA replication; (2) Mutations could be induced by RNA-editing system as a natural defense system from the host; (3) genetic variation from recombination of two viral lineages 8,9,11,12,13. To viruses, mutations could be neutral, which is the majority, harmful or to a less extent represent some advantages to virus [219]. In SARS-CoV-2, the mutations have a lower rate, an estimated 1–2 mutations per month [220], because the RNA polymerase has an unusual proof-reading mechanism [221]. Even so, many mutations have been related to SARS-CoV-2 since the outbreak has started [222,223]. The mutations in SARS-CoV-2 are the most listed in S protein and associated with higher transmissibility and virulence [222,223].
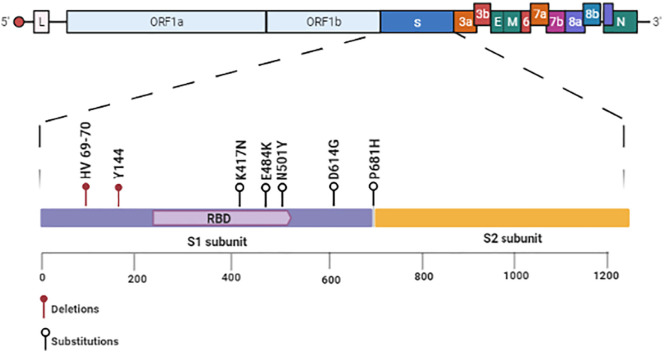
At the beginning of the outbreak, in January of 2020, WHO recognized the circulation of a SARS-CoV-2 variant with a D614G substitution in the S protein (Fig. 7 ). [7,26,109]. According to WHO [7,26,109], the D614G variant was first identified in China and became the dominant variant worldwide by June 2020. The D614G variant, compared to other SARS-CoV-2, has enhancements in infectivity and transmission [222] but does not lead to higher illness.
Fig. 7.
Schematic diagram of mutations in the Spike protein from SARS-CoV-2. All relevant mutations in the Spike protein that lead to the new variants of SARS-CoV-2 are in the RBD. Among those, the most relevant are deletion: HV 69–70 and Y144. Substitutions: K417N, E484K, N501Y (RBD), D614G, and P681H. Created with BioRender.com.
The D614G mutation changes a Gly residue by Asp residue at 614 positions located at the C-terminal region of S1 subunits from S protein, a region directly associated with S2 subunit [222,223]. The advantage of D614G mutation is the high transmission, infectivity, and viral loads in patients with COVID-19 [222,223]. Experiments performed by Plante et al. [224] showed that sera from hamsters infected with the wildtype slightly neutralized the D614G variant. This is a concerning result because it has implications in the vaccine efficacy and thus in antibody therapy.
As happens to other mutations discussed in SARS-CoV-2, the D614G is not in the RBD domain and does not improve the affinity of S protein by ACE2. Indeed, the D614G results in a more stable S protein than wild type S protein and increases the assembly of more functional S protein outside viral particles. Both these advantages increase SARS-CoV-2 infectivity [[222], [223], [224]].
Last December 2020, WHO recognized a new SARS-CoV-2 mutant discovered in the United Kingdom from samples collected 20-Sept-2020 in the county of Kent and another on 21-Sept-2020 from London [225]. This mutant was named SARS-CoV-2 lineage B.1.1.7 (SARS-B117). The mutant SARS-B117 has 17 mutations. Of these, eight mutations were found in the gene responsible for S protein production [225]. The mutations in the S protein were: two deletions: HV 69–70 and Y144 (Fig. 7). The deletions in the 69 and 70 position of the S protein are related to the evasion of S protein from the immune response. Probable, the deletions change the shape of the S protein to make it harder for antibodies to attach. Besides, very recent experiments showed these deletions enable the SARS-B117 to infect cells more efficiently than its wildtype counterpart [225].
The other two mutations in SARS-B117 have biological significance. The Mutation N501Y in RBD (Fig. 7) is said to increase its affinity by ACE2 receptor, increase the transmissibility and infectivity, and make SARS-B117 more contagious. The N501Y mutation changes an amino acid residue of Asn by one of Tyr on the top of each S monomer protein. The region that makes keeps in touch with ACE2 on human cells. In a typical CoV, the top of the S protein is like a fitting puzzle piece. Even though it can catch onto human cells, but the fit is so loose that the virus sometimes falls away and fails to infect the cell. The N501Y mutation came to fix that by making the shape sharp of the puzzle piece, allowing a tighter touch, and enhancing the chance of a successful infection [225]. A new variant of SARS-CoV-2 N501Y·V2 (B.1.351) has emerged in South Africa and is rapidly replacing pre-existing variants.
The other mutation, P681H (Fig. 7), creates a new cleavage site in the intersection of S1 and S2 in the S protein. This cleavage site S1/S2 is unique to SARS-CoV-2, being not present in other related coronaviruses and increase the infection of epithelial cells by SARS-B117 [225]. Therefore, the new virus SARS-B177 has some advantages compared to already circulating SARS-CoV-2 is less immunogenic and has a higher affinity to ACE2 ability to enter within cells. These results revealed the potential and brought concerns about the control of SARS-CoV-2. As of 30 December 2020, the SARS-B117 has spread to 31 other countries proving the high transmissibility of this new variant.
Recently, now in January 2021, the researchers from Oswaldo Cruz Foundation (FIOCRUZ) reported a new variant of SARS-CoV-2, name SARS-CoV-2 B.1.1.28 (SARS-B1128) [226]. The SARS-B1128 was first identified in Amazonas, but there are reports of the same variant, Manaus making a new clade of viruses that emerged in Brazil. The analysis of 69 genomes collected at the hospital from Amazonas states revealed this variant SARS-B1128 still maintains the N501Y mutation and acquired two new mutations K417N and E484K S protein (Fig. 7). Specifically, those mutations are in the RBD portion of the S protein.
By now, there is no scientific or experimental support, but these mutations may have enhanced the ability of SARS-B1128 to bind to ACE2, increasing the transmissibility rates. Based on that, it is feasible to suggest that this high transmissibility is behind the dramatic increase of COVID-19 cases in the state of Amazonas that led to the collapse of the public health system and taken many lives.
Since the beginning of the outbreak, the COVID-19 pandemic has reached every continent in the world. The faster COVID-19 spreads started the run to develop vaccines. Today, some vaccines are already licensed in humans after passing by phase 3 clinical trials. These are the recombinant spike glycoprotein: either mRNA based (the Moderna and Pfizer–BioNTech vaccines), via an adenovirus vector (the Oxford–AstraZeneca, CanSino, and Johnson & Johnson vaccines), or via injection of the protein itself (the Novavax vaccine). The rapid development of vaccines is a breakthrough in the history of vaccines. However, one question arises: Are these vaccines effective against the SARS-CoV-2 new variants?
All the vaccines available today are based on the original SARS-CoV-2 that came up in Wuhan. So, in theory, it is feasible to suggest that variants of SARS-CoV-2 could evolve with resistance to immunity induced by recombinant spike protein vaccines once the mutations lead to a new different type of Spike protein. For instance, a 2020 preprint (non-peer-reviewed manuscript) revealed that a convalescent plasma for hCoV 229E does not neutralize the mutant hCoV 229E leading to individuals less able to neutralize new strains.
From this point, this data suggests that mutations in SARS-CoV-2 could lead to new variants not affected by the vaccine. The length of the spike protein used by licensed vaccines is relatively short (~1270 amino acids), leading to vaccines concentrated only in two sections NTD and RBD domains. Based on that, the antibody response is so focused on some parts of the spike protein. Spike protein mutations might be driven by antigenic drift or even by the selection, either during natural infection or due to the vaccine itself. It is already known that a virus grown under the selective pressure imposed by a single monoclonal antibody targeting a single epitope of a viral protein, mutations in that protein sequence will lead to the loss of neutralization. Additionally, notable showed that SARS-CoV-2 could mutate to escape neutralization by polyclonal antibodies.
Undoubtedly, the spike glycoprotein mutations can affect the efficiency of SARS-CoV-2 neutralization by antibodies, either mono- or polyclonal. Besides the effect in the humoral response, mutations in the spike protein could affect cellular response making the cytotoxic effect of T cells useless. However, only clinical trials with vaccine patients could bring to light this concern.
9. Conclusion
As discussed, all over in this review, the S protein play as a principal actor to SARS-CoV-2 became known and spread worldwide in the last year, causing the third outbreak of coronaviruses. All the new features available to SARS-CoV-2 that led to a higher pathogenesis spectrum, transmissibility, and high affinity to human receptors were enhanced by mutations accumulated by the S protein from SARS-CoV-2 it more efficient than that one from SARS-CoV and MERS-CoV. Even though SARS-CoV and SARS-CoV-2 share similarities, there are differences, which may explain both viruses' completely different behaviors. Most mutations accumulated by S protein from SARS-CoV-2 are hosted in the RBD domain, which is noticeably different from SARS-CoV and MERS-CoV RBD. Here we discuss some mutations that change the amino acid content and atomic location of molecules leading to differences in the S protein from SARS-CoV, MERS-CoV, and SARS-CoV-2. These differences made S protein from SARS-CoV-2 more efficient to bind host receptors than SARS-CoV and MERS-CoV. Mutations also provided an additional cleavage in S1/S2 exclusively from SARS-CoV-2 improving membrane fusion. The unique glycosylation pattern is another advantage accumulated by SARS-CoV-2 S protein playing essential roles in viral tropism, pathogenesis, transmissibility, and evasion of the immune system. Therefore, S protein from pandemic coronaviruses is an essential factor in the show presented by them. Based on that, in-depth studies are required to investigate and understand more about S protein to develop potent drugs and vaccines to eliminate these threats.
CRediT authorship contribution statement
Conceptualization: Pedro F. N. Souza.
Data curation: Pedro F.N. Souza Felipe P. Mesquita, Jackson L. Amaral, Patrícia G. C. Landim, Karollyny R.P. Lima, Marília B. Costa, Izabelle R. Farias, Luina B. Lima.
Formal analysis: Pedro F.N. Souza, Jackson L. Amaral and Felipe P. Mesquita.
Funding acquisition: Pedro F.N. Souza, Felipe P. Mesquita and Raquel C. Montenegro.
Resources: Pedro F.N. Souza, Felipe P. Mesquita and Raquel C. Montenegro.
Supervision: Pedro F.N. Souza.
Writing–original draft: Pedro F.N. Souza, Felipe P. Mesquita, Jackson L. Amaral, Patrícia G. C. Landim, Karollyny R.P. Lima, Marília B. Costa, Izabelle R. Farias, Luina B. Lima.
Writing, review and editing: Pedro F.N. Souza, Felipe P. Mesquita and Raquel C. Montenegro.
Final approve of manuscript and submission: Pedro F.N. Souza.
Ethical approval
Not applicable.
Funding and acknowledgments
Grants from the following Brazilian agencies supported this work: The National Council for Scientific and Technological Development (CNPq), with a doctoral and master grant to JLA and FPM a research grant; the Office to Coordinate Improvement of University Personnel (CAPES) sponsored PFNS with a postdoctoral fellowship.
Declaration of competing interest
All authors declare no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2021.02.203.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Weiss S.R. Forty years with coronaviruses. J. Exp. Med. 2020:217. doi: 10.1084/jem.20200537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman C.M., Frieman M.B. Coronaviruses: important emerging human pathogens. J. Virol. 2014;88:5209–5212. doi: 10.1128/jvi.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ICTV https://talk.ictvonline.org/taxonomy/ Available online: (accessed on Jan 21, 2021)
- 5.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/nejmoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- 7.Coronavirus disease (COVID-19) https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Available online: (accessed on Jan 21, 2021)
- 8.Ding Q., Lu P., Fan Y., Xia Y., Liu M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol. 2020;92:1549–1555. doi: 10.1002/jmv.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X., Cai Y., Huang X., Yu X., Zhao L., Wang F., Li Q., Gu S., Xu T., Li Y., et al. Co-infection with SARS-CoV-2 and influenza a virus in patient with pneumonia, China. Emerg. Infect. Dis. 2020;26:1324–1326. doi: 10.3201/EID2606.200299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khodamoradi Z., Moghadami M., Lotfi M. Co-infection of coronavirus disease 2019 and influenza a: A report from Iran. Arch. Iran. Med. 2020;23:239–243. doi: 10.34172/aim.2020.04. [DOI] [PubMed] [Google Scholar]
- 11.Mahase E. Covid-19: outbreak could last until spring 2021 and see 7.9 million hospitalised in the UK. BMJ. 2020;368 doi: 10.1136/bmj.m1071. [DOI] [PubMed] [Google Scholar]
- 12.Vos L.M., Bruyndonckx R., Zuithoff N.P.A., Little P., Oosterheert J.J., Broekhuizen B.D.L., Lammens C., Loens K., Viveen M., Butler C.C., et al. Lower respiratory tract infection in the community: associations between viral aetiology and illness course. Clin. Microbiol. Infect. 2021;27:96–104. doi: 10.1016/j.cmi.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meo S.A., Alhowikan A.M., Khlaiwi T.A.L., Meo I.M., Halepoto D.M., Iqbal M., Usmani A.M., Hajjar W., Ahmed N. Novel coronavirus 2019-nCoV: prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. Eur. Rev. Med. Pharmacol. Sci. 2020;24:2012–2019. doi: 10.26355/eurrev_202002_20379. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., Yang T., Yu F., Liu J., Liu B., et al. 2020. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I. bioRxiv. 2020.05.24.111823. [DOI] [Google Scholar]
- 15.Shah V.K., Firmal P., Alam A., Ganguly D., Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan Y., Tang F. SARS-CoV-2-mediated immune system activation and potential application in immunotherapy. Med. Res. Rev. 2020;1:1167–1194. doi: 10.1002/med.21756. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., Yang T., Yu F., Liu J., Liu B., et al. 2020. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park M.D. Immune evasion via SARS-CoV-2 ORF8 protein? Nat. Rev. Immunol. 2020;20:408. doi: 10.1038/s41577-020-0360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coronavirus Update (Live) 97,633,202 cases and 2,091,064 deaths from COVID-19 virus pandemic - Worldometer. https://www.worldometers.info/coronavirus/ Available online:
- 20.Wang W., Tang J., Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J. Med. Virol. 2020;92:441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severe Acute Respiratory Syndrome (SARS) https://www.who.int/health-topics/severe-acute-respiratory-syndrome#tab=tab_1 Available online: (accessed on Jan 21, 2021)
- 22.Zhong N.S., Zheng B.J., Li Y.M., Poon L.L.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Memish Z.A., Perlman S., Van Kerkhove M.D., Zumla A. Middle East respiratory syndrome. Lancet. 2020;395:1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Middle East respiratory syndrome coronavirus (MERS-CoV) https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1 Available online: (accessed on Jan 21, 2021) [DOI] [PMC free article] [PubMed]
- 25.Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Eurosurveillance. 2013;18 doi: 10.2807/ese.18.11.20427-en. [DOI] [PubMed] [Google Scholar]
- 26.Coronavirus disease (COVID-19) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available online:
- 27.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijder E.J., Horzinek M.C. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 1993;74:2305–2316. doi: 10.1099/0022-1317-74-11-2305. [DOI] [PubMed] [Google Scholar]
- 30.Enjuanes L., Sola I., Alonso S., Escors D., Zúñiga S. Coronavirus reverse genetics and development of vectors for gene expression. Curr. Top. Microbiol. Immunol. 2005;287:161–197. doi: 10.1007/3-540-26765-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regenmortel M.H.V. van, Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., et al. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the international committee on taxonomy of viruses. Virus Taxon. Classif. Nomencl. viruses. Seventh Rep. Int. Comm. Taxon. Viruses. 2000;1:1123–1128. [Google Scholar]
- 32.Pan X., Ojcius D.M., Gao T., Li Z., Pan C., Pan C. Lessons learned from the 2019-nCoV epidemic on prevention of future infectious diseases. Microbes Infect. 2020;22:86–91. doi: 10.1016/j.micinf.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teeling E.C., Springer M.S., Madsen O., Bates P., O’Brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science (80-. ) 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 34.Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science (80-. ) 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 36.Wang W., Lin X.D., Guo W.P., Zhou R.H., Wang M.R., Wang C.Q., Ge S., Mei S.H., Li M.H., Shi M., et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao Y., Shi M., Chommanard C., Queen K., Zhang J., Markotter W., Kuzmin I.V., Holmes E.C., Tong S. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J. Virol. 2017;91 doi: 10.1128/jvi.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge X.Y., Li J.L., Yang X. Lou, Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacroix A., Duong V., Hul V., San S., Davun H., Omaliss K., Chea S., Hassanin A., Theppangna W., Silithammavong S., et al. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect. Genet. Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science (80-. ) 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 41.Leopardi S., Holmes E.C., Gastaldelli M., Tassoni L., Priori P., Scaravelli D., Zamperin G., De Benedictis P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018;58:279–289. doi: 10.1016/j.meegid.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giannis D., Ziogas I.A., Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao C., Chen W., Zheng S., Zhao J., Wang J., Cao W. Analysis of spatiotemporal characteristics of pandemic SARS spread in mainland China. Biomed. Res. Int. 2016;2016 doi: 10.1155/2016/7247983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12 doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA - J. Am. Med. Assoc. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 46.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alam M.R., Kabir M.R., Reza S. Comorbidities might be a risk factor for the incidence of COVID-19: evidence from a web-based survey. Prev. Med. Rep. 2021;21 doi: 10.1016/j.pmedr.2021.101319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu B., Zeng L.P., Yang X. Lou, Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu B., Ge X., Wang L.F., Shi Z. Bat origin of human coronaviruses coronaviruses: emerging and re-emerging pathogens in humans and animals Susanna Lau positive-strand RNA viruses. Virol. J. 2015:12. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anand S.P., Chen Y., Prévost J., Gasser R., Beaudoin-Bussières G., Abrams C.F., Pazgier M., Finzi A. Interaction of human ACE2 to membrane-bound SARS-CoV-1 and SARS-CoV-2 S glycoproteins. Viruses. 2020;12 doi: 10.3390/v12101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84:11418–11428. doi: 10.1128/jvi.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J. Pathogenicity and transmissibility of 2019-nCoV—A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antibody-Dependent Enhancement of SARS Coronavirus Infection and its Role in the Pathogenesis of SARS | HKMJ Available online: https://www.hkmj.org/abstracts/v22n3Suppl 4/25.htm (accessed on Jan 21, 2021). [PubMed]
- 57.Yoshikawa T., Hill T., Li K., Peters C.J., Tseng C.-T.K. Severe acute respiratory syndrome (SARS) coronavirus-induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte-derived macrophages and dendritic cells. J. Virol. 2009;83:3039–3048. doi: 10.1128/jvi.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sims A.C., Baric R.S., Yount B., Burkett S.E., Collins P.L., Pickles R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting Airways of the Lungs. J. Virol. 2005;79:15511–15524. doi: 10.1128/jvi.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abate S.M., Ahmed Ali S., Mantfardo B., Basu B. Rate of intensive care unit admission and outcomes among patients with coronavirus: a systematic review and meta-analysis. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joynt G.M., Yap H.Y. SARS in the intensive care unit. Curr. Infect. Dis. Rep. 2004;6:228–233. doi: 10.1007/s11908-004-0013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guimarães F. Atuação do fisioterapeuta em unidades de terapia intensiva no contexto da pandemia de COVID-19. Fisioter. em Mov. 2020;33 doi: 10.1590/1980-5918.033.ed01. [DOI] [Google Scholar]
- 62.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. 2020, doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed]
- 64.Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau S.K.P., Wong A.C.P., Lau T.C.K., Woo P.C.Y. Molecular evolution of MERS coronavirus: dromedaries as a recent intermediate host or long-time animal reservoir? Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18102138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Z., Shen L., Gu X. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission. Sci. Rep. 2016;6 doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peiris J.S.M. Medical Microbiology. Eighteenth edition. Elsevier Inc.; 2012. Coronaviruses; pp. 587–593. (ISBN 9780702040894) [Google Scholar]
- 68.Lau S.K.P., Li K.S.M., Luk H.K.H., He Z., Teng J.L.L., Yuen K.-Y., Wernery U., Woo P.C.Y. Middle East respiratory syndrome coronavirus antibodies in Bactrian and hybrid camels from Dubai. mSphere. 2020;5 doi: 10.1128/msphere.00898-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ali M.A., El-Shesheny R., Kandeil A., Shehata M., Elsokary B., Gomaa M., Hassan N., El Sayed A., El-Taweel A., Sobhy H., et al. Cross-sectional surveillance of middle east respiratory syndrome coronavirus (MERS-CoV) in dromedary camels and other mammals in Egypt, august 2015 to January 2016. Eurosurveillance. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.11.30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer B., Müller M.A., Corman V.M., Reusken C.B.E.M., Ritz D., Godeke G.J., Lattwein E., Kallies S., Siemens A., van Beek J., et al. Antibodies against MERS coronavirus in dromedaries, United Arab Emirates, 2003 and 2013. Emerg. Infect. Dis. 2014;20:552–559. doi: 10.3201/eid2004.131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Müller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Bosch B.J., Lattwein E., Hilali M., Musa B.E., et al. Mers coronavirus neutralizing antibodies in camels, eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Said M.Y., Gluecks I., Lattwein E., Bosch B.J., Drexler J.F., et al. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013. Emerg. Infect. Dis. 2014;20:1319–1322. doi: 10.3201/eid2008.140596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cotten M., Watson S.J., Zumla A.I., Makhdoom H.Q., Palser A.L., Ong S.H., Al Rabeeah A.A., Alhakeem R.F., Assiri A., Al-Tawfiq J.A., et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. 2014;5 doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Letko M., Miazgowicz K., McMinn R., Seifert S.N., Sola I., Enjuanes L., Carmody A., van Doremalen N., Munster V. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 2018;24:1730–1737. doi: 10.1016/j.celrep.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coleman C.M., Matthews K.L., Goicochea L., Matthew M.B. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J. Gen. Virol. 2014;95:408–412. doi: 10.1128/jvi.00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tai W., Wang Y., Fett C.A., Zhao G., Li F., Perlman S., Jiang S., Zhou Y., Du L. Recombinant receptor-binding domains of multiple Middle East respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J. Virol. 2017;91 doi: 10.1128/jvi.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Douglas M.G., Kocher J.F., Scobey T., Baric R.S., Cockrell A.S. Adaptive evolution influences the infectious dose of MERS-CoV necessary to achieve severe respiratory disease. Virology. 2018;517:98–107. doi: 10.1016/j.virol.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y., Rajashankar K.R., Yang Y., Agnihothram S.S., Liu C., Lin Y.-L., Baric R.S., Li F. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:10777–10783. doi: 10.1128/jvi.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Du L., Zhao G., Kou Z., Ma C., Sun S., Poon V.K.M., Lu L., Wang L., Debnath A.K., Zheng B.-J., et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013;87:9939–9942. doi: 10.1128/jvi.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mou H., Raj V.S., van Kuppeveld F.J.M., Rottier P.J.M., Haagmans B.L., Bosch B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87:9379–9383. doi: 10.1128/jvi.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/jvi.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peck K.M., Scobey T., Swanstrom J., Jensen K.L., Burch C.L., Baric R.S., Heise M.T. Permissivity of dipeptidyl peptidase 4 orthologs to Middle East respiratory syndrome coronavirus is governed by glycosylation and other complex determinants. J. Virol. 2017;91 doi: 10.1128/jvi.00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peck K.M., Cockrell A.S., Yount B.L., Scobey T., Baric R.S., Heise M.T. Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection. J. Virol. 2015;89:4696–4699. doi: 10.1128/jvi.03445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cockrell A.S., Peck K.M., Yount B.L., Agnihothram S.S., Scobey T., Curnes N.R., Baric R.S., Heise M.T. Mouse Dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88:5195–5199. doi: 10.1128/jvi.03764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barlan A., Zhao J., Sarkar M.K., Li K., McCray P.B., Perlman S., Gallagher T. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88:4953–4961. doi: 10.1128/jvi.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arabi Y.M., Al-Omari A., Mandourah Y., Al-Hameed F., Sindi A.A., Alraddadi B., Shalhoub S., Almotairi A., Al Khatib K., Abdulmomen A., et al. Critically ill patients with the middle east respiratory syndrome: a multicenter retrospective cohort study. Crit. Care Med. 2017;45:1683–1695. doi: 10.1097/CCM.0000000000002621. [DOI] [PubMed] [Google Scholar]
- 91.Arabi Y.M., Arifi A.A., Balkhy H.H., Najm H., Aldawood A.S., Ghabashi A., Hawa H., Alothman A., Khaldi A., Al Raiy B. Clinical course and outcomes of critically ill patients with middle east respiratory syndrome coronavirus infection. Ann. Intern. Med. 2014;160 doi: 10.7326/m13-2486. [DOI] [PubMed] [Google Scholar]
- 92.Al-Osail A.M., Al-Wazzah M.J. The history and epidemiology of Middle East respiratory syndrome corona virus. Multidiscip. Respir. Med. 2017;12 doi: 10.1186/s40248-017-0101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Azhar E.I., Lanini S., Ippolito G., Zumla A. Advances in Experimental Medicine and Biology. vol. 972. Springer; New York LLC: 2017. The middle east respiratory syndrome coronavirus – a continuing risk to global health security; pp. 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Al-Dorzi H.M., Alsolamy S., Arabi Y.M. Critically ill patients with Middle East respiratory syndrome coronavirus infection. Crit. Care. 2016;20 doi: 10.1186/s13054-016-1234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kupferschmidt K. Camel vaccine offers hope to stop MERS: vaccinated animals shed less virus, but is that good enough to prevent human outbreaks? Science (80-. ). 2015;350:1453. doi: 10.1126/science.350.6267.1453. [DOI] [PubMed] [Google Scholar]
- 98.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Drosten C., Meyer B., Müller M.A., Corman V.M., Al-Masri M., Hossain R., Madani H., Sieberg A., Bosch B.J., Lattwein E., et al. Transmission of MERS-coronavirus in household contacts. N. Engl. J. Med. 2014;371:828–835. doi: 10.1056/nejmoa1405858. [DOI] [PubMed] [Google Scholar]
- 100.Oboho I.K., Tomczyk S.M., Al-Asmari A.M., Banjar A.A., Al-Mugti H., Aloraini M.S., Alkhaldi K.Z., Almohammadi E.L., Alraddadi B.M., Gerber S.I., et al. 2014 MERS-CoV outbreak in Jeddah — a link to health care facilities. N. Engl. J. Med. 2015;372:846–854. doi: 10.1056/nejmoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Senga M., Arabi Y.M., Fowler R.A. Clinical spectrum of the Middle East respiratory syndrome coronavirus (MERS-CoV) J. Infect. Public Health. 2017;10:191–194. doi: 10.1016/j.jiph.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alghamdi M., Mushtaq F., Awn N., Shalhoub S. MERS CoV infection in two renal transplant recipients: case report. Am. J. Transplant. 2015;15:1101–1104. doi: 10.1111/ajt.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alsaad K.O., Hajeer A.H., Al Balwi M., Al Moaiqel M., Al Oudah N., Al Ajlan A., AlJohani S., Alsolamy S., Gmati G.E., Balkhy H., et al. Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection – clinicopathological and ultrastructural study. Histopathology. 2018;72:516–524. doi: 10.1111/his.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cha R.H., Joh J.S., Jeong I., Lee J.Y., Shin H.S., Kim G., Kim Y. Renal complications and their prognosis in Korean patients with Middle East respiratory syndrome-coronavirus from the central MERS-CoV designated hospital. J. Korean Med. Sci. 2015;30:1807–1814. doi: 10.3346/jkms.2015.30.12.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ng D.L., Al Hosani F., Keating M.K., Gerber S.I., Jones T.L., Metcalfe M.G., Tong S., Tao Y., Alami N.N., Haynes L.M., et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of middle east respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am. J. Pathol. 2016;186:652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A.T., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369:407–416. doi: 10.1056/nejmoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.WHO Coronavirus Disease (COVID-19) Dashboard | WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/ Available online:
- 110.Bassetti M., Vena A., Giacobbe D.R. The novel Chinese coronavirus (2019-nCoV) infections: challenges for fighting the storm. Eur. J. Clin. Investig. 2020:50. doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harrison A.G., Lin T., Wang P., et al. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends. Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou P., Yang X. Lou, Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hemida M.G., Ba Abduallah M.M. The SARS-CoV-2 outbreak from a one health perspective. One Heal. 2020;10 doi: 10.1016/j.onehlt.2020.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:127–147. doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang T., Wu Q., Zhang Z. 2020. Pangolin Homology Associated with 2019-nCoV. bioRxiv. 2020.02.19.950253. [DOI] [Google Scholar]
- 120.Nao N., Yamagishi J., Miyamoto H., Igarashi M., Manzoor R., Ohnuma A., Tsuda Y., Furuyama W., Shigeno A., Kajihara M., et al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. MBio. 2017:8. doi: 10.1128/mBio.02298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rastogi Y.R., Sharma A., Nagraik R., Aygün A., Şen F. The novel coronavirus 2019-nCoV: its evolution and transmission into humans causing global COVID-19 pandemic. Nat. Rev. Microbiol. 2020;26:1–8. doi: 10.1007/s13762-020-02781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sabioni L., De Lorenzo A., Lamas C., Muccillo F., Castro-Faria-Neto H.C., Estato V., Tibirica E. Systemic microvascular endothelial dysfunction and disease severity in COVID-19 patients: evaluation by laser Doppler perfusion monitoring and cytokine/chemokine analysis. Microvasc. Res. 2021;134 doi: 10.1016/j.mvr.2020.104119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dong M., Zhang J., Ma X., Tan J., Chen L., Liu S., Xin Y., Zhuang L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/nejmc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 129.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lomniczi B. Biological properties of avian coronavirus RNA. J. Gen. Virol. 1977;36:531–533. doi: 10.1099/0022-1317-36-3-531. [DOI] [PubMed] [Google Scholar]
- 131.Lai M.M., Liao C.L., Lin Y.J., Zhang X. Coronavirus: how a large RNA viral genome is replicated and transcribed. Infect. Agents Dis. 1994;3:98–105. [PubMed] [Google Scholar]
- 132.de Haan C.A.M., Masters P.S., Shen X., Weiss S., Rottier P.J.M. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Norman J.O., McClurkin A.W., Bachrach H.L. Infectious nucleic acid from a transmissible agent causing gastroenteritis in pigs. J. Comp. Pathol. 1968;78:227–235. doi: 10.1016/0021-9975(68)90099-6. [DOI] [PubMed] [Google Scholar]
- 134.Schochetman G., Stevens R.H., Simpson R.W. Presence of infectious polyadenylated RNA in coronavirus avian bronchitis virus. Virology. 1977;77:772–782. doi: 10.1016/0042-6822(77)90498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wege H., Mueller A., Ter Meulen V. Genomic RNA of the murine coronavirus JHM. J. Gen. Virol. 1978;41:217–227. doi: 10.1099/0022-1317-41-2-217. [DOI] [PubMed] [Google Scholar]
- 136.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/nejmoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dinman J.D. Programmed ribosomal frameshifting goes beyond viruses. Microbe. 2006;1:521–527. doi: 10.1128/microbe.1.521.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jacobs J.L., Belew A.T., Rakauskaite R., Dinman J.D. Identification of functional, endogenous programmed - 1 ribosomal frameshift signals in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:165–174. doi: 10.1093/nar/gkl1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Klobutcher L.A., Farabaugh P.J. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell. 2002;111:763–766. doi: 10.1016/s0092-8674(02)01138-8. [DOI] [PubMed] [Google Scholar]
- 140.Hammell A.B., Taylor R.C., Peltz S.W., Dinman J.D. Identification of putative programmed −1 ribosomal frameshift signals in large DNA databases. Genome Res. 1999;9:417–427. doi: 10.1101/gr.9.5.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 142.Cobucci-Ponzano B., Rossi M., Moracci M. Translational recoding in archaea. Extremophiles. 2012;16:793–803. doi: 10.1007/s00792-012-0482-8. [DOI] [PubMed] [Google Scholar]
- 143.Baric R.S., Yount B. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dalton A., Haguenau F. 1973. Ultrastructure of Animal Viruses and Bacteriophages : An Atlas. [Google Scholar]
- 145.Davies H.A., Macnaughton M.R. Comparison of the morphology of three coronaviruses. Arch. Virol. 1979;59:25–33. doi: 10.1007/BF01317891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ng M.L., Lee J.W.M., Leong M.L.N., Ling A.E., Tan H.C., Ooi E.E. Topographic changes in SARS coronavirus-infected cells during late stages of infection. Emerg. Infect. Dis. 2004;10:1907–1914. doi: 10.3201/eid1011.040195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wüthrich K., et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bárcena M., Oostergetel G.T., Bartelink W., Faas F.G.A., Verkleij A., Rottier P.J.M., Koster A.J., Bosch B.J. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. U. S. A. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.de Haan C.A.M., Kuo L., Masters P.S., Vennema H., Rottier P.J.M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Locker J.K., Griffiths G., Horzinek M.C., Rottier P.J. O-glycosylation of the coronavirus M protein. Differential localization of sialyltransferases in N- and O-linked glycosylation. J. Biol. Chem. 1992;267:14094–14101. doi: 10.1016/S0021-9258(19)49683-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Escors D., Camafeita E., Ortego J., Laude H., Enjuanes L. Organization of two Transmissible Gastroenteritis Coronavirus Membrane Protein Topologies within the Virion and Core. J. Virol. 2001;75:12228–12240. doi: 10.1128/jvi.75.24.12228-12240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76:4987–4999. doi: 10.1128/jvi.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.de Haan C.A.M., Vennema H., Rottier P.J.M. Assembly of the coronavirus envelope: Homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nieto-Torres J.L., DeDiego M.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/jvi.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Curtis K.M., Yount B., Baric R.S. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 2002;76:1422–1434. doi: 10.1128/jvi.76.3.1422-1434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.DeDiego M.L., Alvarez E., Almazán F., Rejas M.T., Lamirande E., Roberts A., Shieh W.-J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kuo L., Hurst K.R., Masters P.S. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 2007;81:2249–2262. doi: 10.1128/JVI.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nieto-Torres J.L., DeDiego M.L., Álvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li Y., Surya W., Claudine S., Torres J. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem. 2014;289:12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., Dediego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta. 2013;1828:2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Torres J., Maheswari U., Parthasarathy K., Ng L., Liu D.X., Gong X. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16:2065–2071. doi: 10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Laude H., Rasschaert D., Huet J.C. Sequence and N-terminal processing of the transmembrane protein E1 of the coronavirus transmissible gastroenterits virus. J. Gen. Virol. 1987;68:1687–1693. doi: 10.1099/0022-1317-68-6-1687. [DOI] [PubMed] [Google Scholar]
- 170.Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus Nucleocapsid protein. J. Virol. 2005;79:13285–13297. doi: 10.1128/jvi.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Narayanan K., Chen C.-J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/jvi.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen H., Gill A., Dove B.K., Emmett S.R., Kemp C.F., Ritchie M.A., Dee M., Hiscox J.A. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J. Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zakhartchouk A.N., Viswanathan S., Mahony J.B., Gauldie J., Babiuk L.A. Severe acute respiratory syndrome coronavirus nucleocapsid protein expressed by an adenovirus vector is phosphorylated and immunogenic in mice. J. Gen. Virol. 2005;86:211–215. doi: 10.1099/vir.0.80530-0. [DOI] [PubMed] [Google Scholar]
- 174.Song H.C., Seo M.-Y., Stadler K., Yoo B.J., Choo Q.-L., Coates S.R., Uematsu Y., Harada T., Greer C.E., Polo J.M., et al. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2004;78:10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S., Stroeher U., Bastien N., Dasuri K.V.N., Cheng K., et al. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shang J., Yushun W., Chuming L., Gang Y., Qibin G., Ashley A., Fang L., et al. Cell entry mechanisms of SARS-CoV-2. PNAS. 2020;117:11727–11734. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Du L., Kao R.Y., Zhou Y., He Y., Zhao G., Wong C., Jiang S., Yuen K.-Y., Jin D.-Y., Zheng B.-J. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 2007;359:174–179. doi: 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 184.Souza P.F.N., Lopes F.E.S., Amaral J.L., Freitas C.D.T., Oliveira J.T.A. International journal of biological macromolecules a molecular docking study revealed that synthetic peptides induced conformational changes in the structure of SARS-CoV-2 spike glycoprotein , disrupting the interaction with human ACE2 receptor. Int. J. Biol. Macromol. 2020;164:66–76. doi: 10.1016/j.ijbiomac.2020.07.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Souza P.F.N., Amaral J.L., Bezerra L.P., Lopes F.E.S., Freire V.N., Oliveira J.T.A., Freitas C.D.T. ACE2-derived peptides interact with the RBD domain of SARS-CoV-2 spike glycoprotein, disrupting the interaction with the human ACE2 receptor. J. Biomol. Struct. Dyn. 2021:1–14. doi: 10.1080/07391102.2020.1871415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chambers, P.; Pringle, C.R.; Easton, A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990, 71 ( Pt 12, 3075–3080, doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed]
- 188.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet (London, England) 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T.K., Wang Q., Du L., Tan W., Wilson I.A., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Widagdo W., Okba N.M.A., Li W., de Jong A., de Swart R.L., Begeman L., van den Brand J.M.A., Bosch B.-J., Haagmans B.L. Species-specific Colocalization of Middle East respiratory syndrome coronavirus attachment and entry receptors. J. Virol. 2019;93 doi: 10.1128/jvi.00107-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Weissenhorn W., Dessen A., Calder L.J., Harrison S.C., Skehel J.J., Wiley D.C. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 192.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., Wei D., Zhang Y., Sun X.X., Gong L., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020;5:1–10. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li Y., Xu J., Chen L., Zhong W. De, Zhang Z., Mi L., Zhang Y., Liao C.G., Bian H.J., Jiang J.L., et al. HAb18G (CD147), a cancer-associated biomarker and its role in cancer detection. Histopathology. 2009;54:677–687. doi: 10.1111/j.1365-2559.2009.03280.x. [DOI] [PubMed] [Google Scholar]
- 198.Pushkarsky T., Zybarth G., Dubrovsky L., Yurchenko V., Tang H., Guo H., Toole B., Sherry B., Bukrinsky M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin a. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., Sun Z., Liu K., Shan C., Lin A., Jiang S., Xie Y., Zhou Q., Lu L., Huang J., Li X. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B., et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., Zhang Y., Fan C., Li D., Deng Y., Sun J., Gong J., Yang X., Wang Y., Wang X., Li J., Yang H., Li H., Zhang Z., Wang R., Du P., Zong Y., Yin F., Zhang W., Wang N., Peng Y., Lin H., Feng J., Qin C., Chen W., Gao Q., Zhang R., Cao Y., Zhong H. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 203.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Österlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (80-. ) 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Daly J.L., Simonetti B., Antón-Plágaro C., Williamson M.K., Shoemark D.K., Simón-Gracia L., Klein K., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Cullen P.J., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. BioRxiv. 2020;865:861–865. doi: 10.1101/2020.06.05.134114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tang X., Yang M., Duan Z., Liao Z., Liu L., Cheng R., Fang M., Wang G., Liu H., Xu J., Kamau P.M., Zhang Z., Yang L., Zhao X., Peng X., Lai R. Transferrin receptor is another receptor for SARS-CoV-2 entry. BioRxiv. 2020;20 http://biorxiv.org/content/early/2020/10/23/2020.10.23.350348.abstract 2020.10.23.350348. [Google Scholar]
- 206.R. Amraei, W. Yin, M.A. Napoleon, E.L. Suder, J. Berrigan, Q. Zhao, J. Olejnik, K.B. Chandler, C. Xia, J. Feldman, B.M. Hauser, T.M. Caradonna, A.G. Schmidt, S. Gummuluru, E. Mühlberger, V. Chitalia, ,6,7 , (2020).
- 207.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Souza P.F.N., Carvalho F.E.L. Killing two birds with one stone: how do Plant viruses break down plant defenses and manipulate cellular processes to replicate themselves? J. Plant Biol. 2019;62:170–180. doi: 10.1007/s12374-019-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fan Y., Zhao K., Shi Z.L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11 doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Souza P.F.N. Masters of manipulation: how do positive-sense RNA viruses employ plant proteins to replicate, move from cell to cell, and overcome antiviral immunity? J. Plant Dis. Prot. 2020 doi: 10.1007/s41348-020-00342-w. [DOI] [Google Scholar]
- 212.Brufsky A. Distinct viral clades of SARS-CoV-2: implications for modeling of viral spread. J. Med. Virol. 2020;92:1386–1390. doi: 10.1002/jmv.25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Spinello A., Saltalamacchia A., Magistrato A. Is the rigidity of SARS-CoV-2 spike receptor-binding motif the Hallmark for its enhanced infectivity? Insights from all-atom simulations. J. Phys. Chem. Lett. 2020;11:4785–4790. doi: 10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- 214.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chang T.-J., Yang D.-M., Wang M.-L., Liang K.-H., Tsai P.-H., Chiou S.-H., Lin T.-H., Wang C.-T. Genomic analysis and comparative multiple sequences of SARS-CoV2. J. Chin. Med. Assoc. 2020;83:537–543. doi: 10.1097/JCMA.0000000000000335. [DOI] [PubMed] [Google Scholar]
- 216.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39 doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/jvi.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kimura M., Ota T. On the rate of molecular evolution. J. Mol. Evol. 1971;1:1–17. doi: 10.1007/BF01659390. [DOI] [PubMed] [Google Scholar]
- 220.Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6 doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang L., Jackson C., Mou H., Ojha A., Rangarajan E., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- 223.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020:1–6. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations - SARS-CoV-2 coronavirus / nCoV-2019 genomic epidemiology - Virological. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 Available online:
- 226.Fiocruz publica Nota Técnica sobre nova variante do Sars-CoV-2 no Amazonas. https://portal.fiocruz.br/noticia/fiocruz-publica-nota-tecnica-sobre-nova-variante-do-sars-cov-2-no-amazonas Available online: (accessed on Jan 22, 2021)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures