Figure 1.
Single-cell RNA-seq (scRNA-seq) analysis revealed region-specific microglial subtypes from the two-month-old wild-type (Wt) brain cortex (2mWtBr) and spinal cord (2mWtSp)
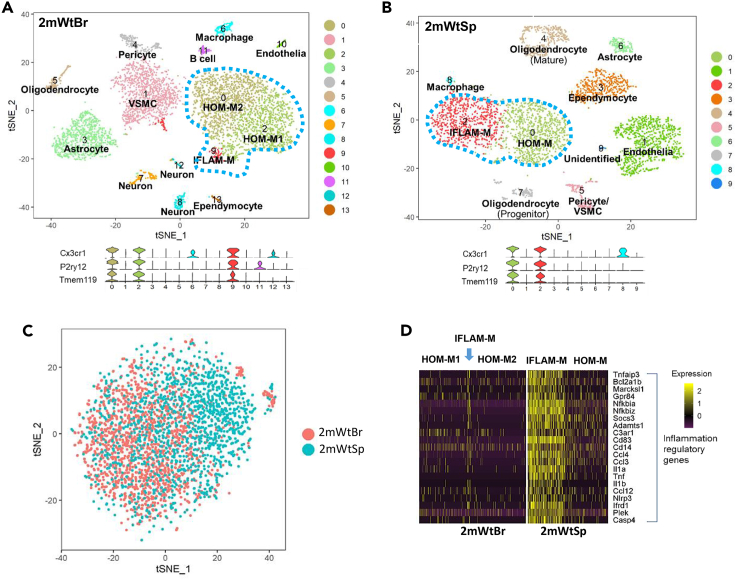
(A and B) t-SNE plots identified microglial clusters (blue dot line circled) in 2mWtBr (A) and 2mWtSp (B). Based on the expressed signature genes, the microglial clusters were defined as homeostatic microglia (HOM-M) and proinflammatory microglia (IFLAM-M). The cortical HOM-M were further segregated into HOM-M1 and HOM-M2 groups. Violin plots below the t-SNE plots show the expression of the microglial markers Cx3cr1, P2ry12, and Tmem119 from the t-SNE clusters. We defined the clusters with detectable expression of all three markers as microglia. VSMC: vascular smooth muscle cell.
(C) Seurat's canonical correlation analysis (CCA) showed that spinal microglia partially overlapped with cortical microglia, indicating cortical and spinal specific microglial heterogeneity.
(D) Gene expression heatmap shows the population of inflammatory microglia was big in the spinal cord but small in the cortex. The columns in D represent the subset of all microglia from 2mWtBr and 2mWtSp. Each row: a gene; each column: a microglial cell. See also Figures S1 and S2.