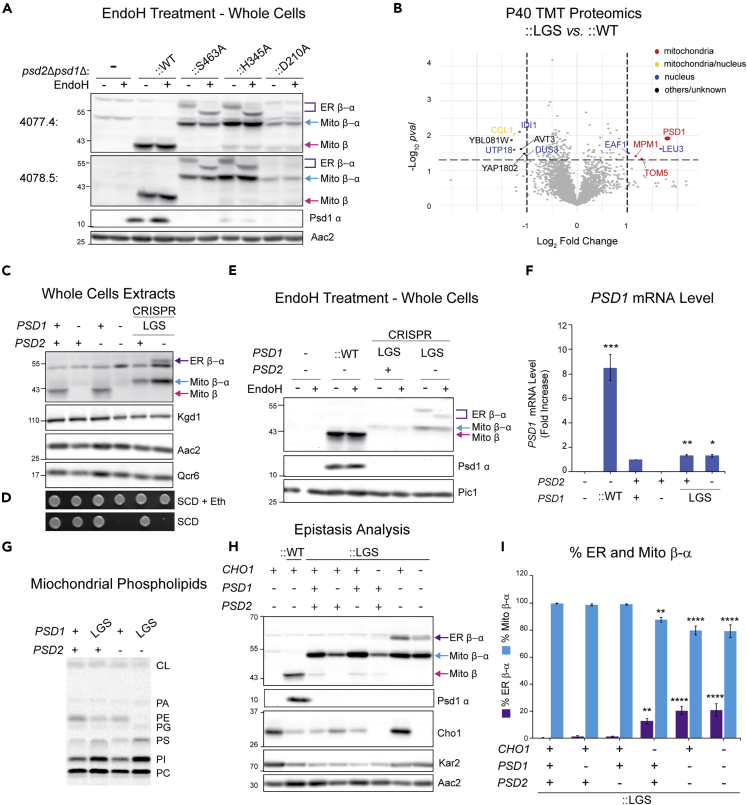
Figure 3.
Glycosylation of endogenous and overexpressed mutant, nonfunctional Psd1 requires severe disruption of cellular PE metabolism
(A) Cell extracts from the indicated strains grown at 30°C in YPD were treated with EndoH as listed and analyzed by immunoblot using the designated anti-Psd1 antisera; Aac2 served as loading control. ER β-α, glycosylated mutant Psd1; Mito β-α, mutant Psd1 not glycosylated (n = 3).
(B) TMT comparison of ER (P40) proteomes from psd2Δpsd1Δ::LGS (n = 3 preps) and psd2Δpsd1Δ::WT yeast (n = 4 preps). LGS; autocatalytic mutant Psd1.
(C) Cell extracts from the indicated strains grown at 30°C in YPD were immunoblotted for the Psd1 β subunit; Kgd1, Aac2, and Qcr6 served as loading controls. LGS was knocked into endogenous PSD1 of the listed strains via Hi-CRISPR (n = 3).
(D) Serial dilutions of same strains as in (C) were spotted onto SCD with or without 2 mM ethanolamine (+Eth) and incubated at 30°C for 3 days (n = 3).
(E) EndoH sensitivity assay of cell extracts from indicated strains grown in YPD at 30°C. Immunoblots were probed for Psd1 β (4,077.4) and Psd1 α (FLAG); Pic1 served as loading control (n = 3).
(F) PSD1 mRNA level in the indicated strains determined by two-step reverse transcription-quantitative PCR of PSD1 and normalized to ACT1 (means ± SEM, n = 4). Statistical differences (1 symbol, p ≤ 0.05; 2 symbols p ≤ 0.01; 3 symbols p ≤ 0.001 compared WT (PSD2 +, PSD1 +) were calculated with unpaired Student's t-test.
(G) Mitochondrial phospholipids from the indicated strains were labeled overnight in YPD medium spiked with 14C-Acetate and separated by TLC (n = 6).
(H) Cell extracts derived from the indicated strains grown at 30°C in YPD were analyzed by immunoblot for Psd1 β (4,077.4), Psd1 α (FLAG), Cho1 (PS synthase), and Kar2 (ER chaperone); Aac2 acted as loading control (n = 5).
(I) The relative abundance of ER β-α and Mito β-α in LGS mutant Psd1 expressing yeast of the indicated genotype was determined (means ± SEM for n = 5). Statistical differences (2 symbols p ≤ 0.01; 4 symbols p ≤ 0.0001) were determined by one-way analysis of variance (ANOVA) with Dunnett's multiple comparison test.