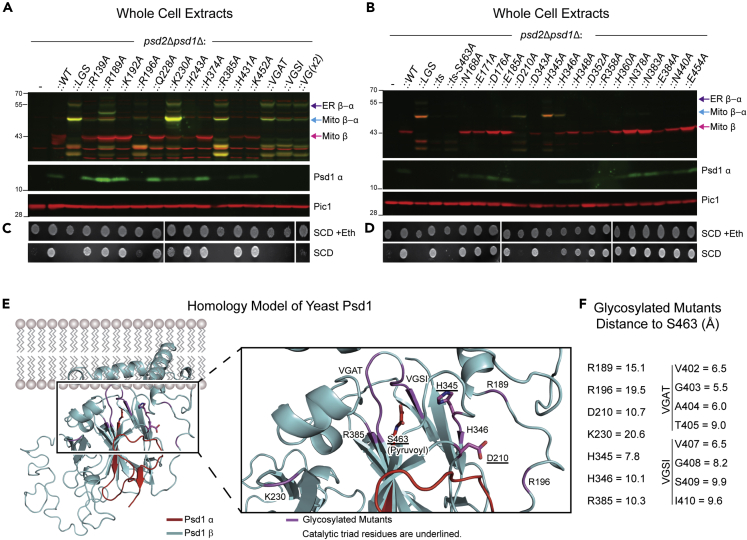
Figure 4.
Mutations that indirectly impair autocatalysis are also glycosylated
(A and B) Cell extracts derived from the indicated strains grown at 30°C in YPD were analyzed by fluorescent immunoblot for Psd1 β (red) and Psd1 α (green); Pic1 acted as loading control (n = 3).
(C and D) The same strains in (A and B) were pre-cultured at 30°C in YPD, spotted onto SCD with (+) or without (−) 2 mM ethanolamine and incubated at 30°C for 3 days (n = 3).
(E) Homology model structure of Psd1 based on the E. coli PSD structure (PDB code: 6L06)—view from the side of membrane. The α subunit and β subunit are colored in red and cyan, respectively. The right panel is a magnified view around S463 (pyruvoyl group at N-terminus of α subunit). Residues whose mutation to alanine impairs autocatalysis are indicated and colored in magenta. Autocatalytic triad residues are underlined and shown in stick form.
(F) The distances were measured between the carbonyl carbon of the pyruvoyl group (S463) and the alpha carbon of the residues whose mutation to alanine impairs autocatalysis.