Figure 7.
Supplements that diminish amount of glycosylated mutant Psd1 act by distinct mechanisms
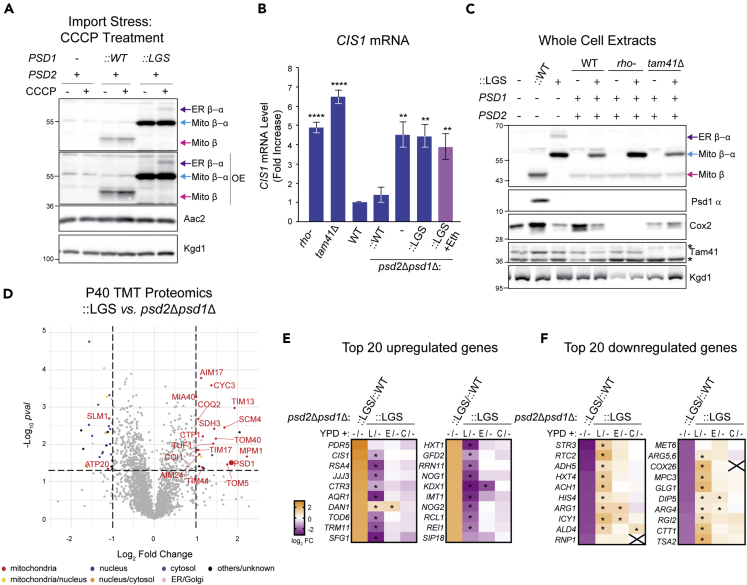
(A) The indicated strains were cultured overnight at 30°C in YPD without or with 10 μM CCCP. Cell extracts were harvested and immunoblotted as designated. OE indicates overexposed blot in which the Mito β-α signals are saturated. (n = 3).
(B) CIS1 mRNA levels in the indicated strains grown in YPD at 30°C were determined by two-step reverse transcription-quantitative PCR and normalized to ACT1 (mean ± SEM, n = 5). Statistical differences (2 symbols p ≤ 0.01; 4 symbols p ≤ 0.0001) versus WT were calculated with unpaired Student's t-test.
(C) Cell extracts from the indicated strains, untransformed or transformed with the autocatalytic LGS Psd1 mutant, grown overnight in YPD were immunoblotted for Psd1 β (4,077.4), Psd1 α (FLAG), Cox2 (encoded by mtDNA), and Tam41; Kgd1 acted as loading control (n = 3). ∗, nonspecific bands.
(D) TMT comparison of ER (P40) proteomes from psd2Δpsd1Δ::LGS and psd2Δpsd1Δ yeast (n = 3 preps each). LGS; autocatalytic mutant Psd1.
(E and F) Heatmaps of gene expression analysis from RNAseq with the indicated strains grown in YPD medium alone or YPD supplemented with lyso-PE (L), ethanolamine (E), or choline (C). padj values that were significantly different from YPD medium alone (p ≤ 0.05) are designated (∗). (E) The top 20 upregulated genes in ::LGS vs ::WT and how each supplement does or does not affect their expression relative to ::LGS grown in YPD alone. (F) The top 20 downregulated genes in ::LGS vs ::WT and how each supplement does or does not affect their expression relative to ::LGS grown in YPD alone.