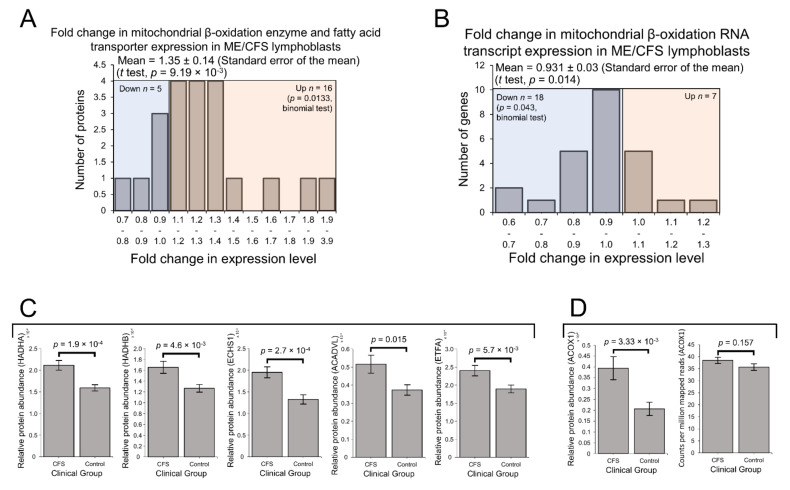
Figure 5.
Expression of proteins involved in mitochondrial and peroxisomal fatty acid β-oxidation was elevated in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) lymphoblasts. Error bars represent the standard error of the mean. RNA sequencing transcriptomics experiment: ME/CFS n = 23, control n = 17. Each cell line was sampled once. Mass spectrometry proteomics experiment: ME/CFS n = 34, control n = 31. Each cell line was sampled once, or twice for a subset of healthy controls arbitrarily selected to act an internal control between experiments in the proteomics work. (A) A total of 21 proteins involved in mitochondrial fatty acid β-oxidation and transport were detected within the whole-cell proteomes of ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given protein in the CFS group divided by the mean abundance in the control group. The proportion of detected proteins that were upregulated (binomial test with Ho set to 0.5 and H1 being that the upregulated proportion was greater) and the average extent of the upregulation (single-sample t test with Ho ≤ m1 and H1 m>1) were statistically significant. (B) A total of 25 RNA transcripts encoding proteins involved in mitochondrial fatty acid β-oxidation and transport were detected by RNA sequencing within the whole-cell transcriptomes of ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given transcript in the CFS group divided by the mean abundance in the control group. The proportions of reduced or elevated transcripts were not significantly different (binomial test with Ho p = 0.5) nor was the average magnitude of expression (single-sample t test with Ho m = 1). (C) The expression levels of both subunits of the mitochondrial trifunctional enzyme complex Hydroxyacyl-CoA dehydrogenase/3-keotacyl-CoA thiolase (HADHA and HADHB), very long-chain specific acyl-CoA dehydrogenase (ACADVL), enoyl-CoA hydratase (ECHS1) and electron transfer flavoprotein subunit alpha (ETFA (are significantly elevated in whole-cell mass spectrometry proteomics experiments (independent t test). Relative abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments. (D) The expression of Acyl-CoA oxidase 1 (ACOX1) was significantly elevated in whole-cell mass spectrometry proteomics experiments (independent t test). Relative abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments. ACOX1 expression was not altered at the transcriptional level as measured by whole-cell RNA sequencing transcriptomics. Counts per million mapped reads were calculated for each gene transcript.